Amplicon-Metagenomic Analysis of Fungi from Antarctic Terrestrial Habitats
- PMID: 29184546
- PMCID: PMC5694453
- DOI: 10.3389/fmicb.2017.02235
Amplicon-Metagenomic Analysis of Fungi from Antarctic Terrestrial Habitats
Abstract
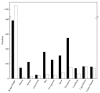
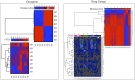
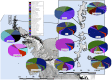
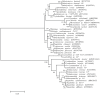
In cold environments such as polar regions, microorganisms play important ecological roles, and most of our knowledge about them comes from studies of cultivable microorganisms. Metagenomic technologies are powerful tools that can give a more comprehensive assessment of microbial communities, and the amplification of rDNA followed by next-generation sequencing has given good results in studies aimed particularly at environmental microorganisms. Culture-independent studies of microbiota in terrestrial habitats of Antarctica, which is considered the driest, coldest climate on Earth, are increasing and indicate that micro-diversity is much higher than previously thought. In this work, the microbial diversity of terrestrial habitats including eight islands of the South Shetland Archipelago, two islands on the Antarctic Peninsula and Union Glacier, was studied by amplicon-metagenome analysis. Molecular analysis of the studied localities clustered together the islands of the South Shetland Archipelago, except Greenwich Island, and separated them from the Litchfield and Lagotellerie islands and Union Glacier, which is in agreement with the latitudinal difference among them. Among fungi, 87 genera and 123 species were found, of which species belonging to 37 fungal genera not previously cultivated from Antarctica were detected. Phylogenetic analysis, including the closest BLAST-hit sequences, clustered fungi in 11 classes being the most represented Lecanoromycetes and Eurotiomycetes.
Keywords: Antarctic diatoms; Antarctic fungi; Antarctic microeukaryotes; amplicon-metagenome; cold-terrestrial microeukaryotes.
Figures





References
-
- Arenz B. E., Blanchette R. A., Farrell R. L. (2014). “Fungal diversity in Antarctic soils,” in Antarctic Terrestrial Microbiology, ed. Cowan D. (Berlin: Springer; ), 35–53.
-
- Bartasun P., Cieśliński H., Kur J. (2012). An examination of an Antarctic soil metagenomic-derivate putative methylthioadenosine phosphorylase gene as a novel reporter gene for promoter trapping. J. Gen. Appl. Microbiol. 58 387–395. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

