Consensus queries in ligand-based virtual screening experiments
- PMID: 29185065
- PMCID: PMC5705545
- DOI: 10.1186/s13321-017-0248-5
Consensus queries in ligand-based virtual screening experiments
Abstract
Background: In ligand-based virtual screening experiments, a known active ligand is used in similarity searches to find putative active compounds for the same protein target. When there are several known active molecules, screening using all of them is more powerful than screening using a single ligand. A consensus query can be created by either screening serially with different ligands before merging the obtained similarity scores, or by combining the molecular descriptors (i.e. chemical fingerprints) of those ligands.
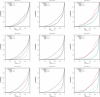
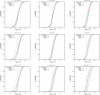
Results: We report on the discriminative power and speed of several consensus methods, on two datasets only made of experimentally verified molecules. The two datasets contain a total of 19 protein targets, 3776 known active and ~ 2 × 106 inactive molecules. Three chemical fingerprints are investigated: MACCS 166 bits, ECFP4 2048 bits and an unfolded version of MOLPRINT2D. Four different consensus policies and five consensus sizes were benchmarked.
Conclusions: The best consensus method is to rank candidate molecules using the maximum score obtained by each candidate molecule versus all known actives. When the number of actives used is small, the same screening performance can be approached by a consensus fingerprint. However, if the computational exploration of the chemical space is limited by speed (i.e. throughput), a consensus fingerprint allows to outperform this consensus of scores.
Keywords: Chemical fingerprint; Consensus query; ECFP4; Ligand-based virtual screening (LBVS); MACCS; MOLPRINT2D; Potency scaling; Several bioactives; Similarity search; Tanimoto score.
Figures







References
-
- Johnson MA, Maggiora GM. Concepts and applications of molecular similarity. New York: Wiley; 1990.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

