RAS ubiquitylation modulates effector interactions
- PMID: 29185849
- PMCID: PMC7549706
- DOI: 10.1080/21541248.2017.1371267
RAS ubiquitylation modulates effector interactions
Abstract
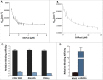
RAS proteins function as molecular switches that regulate cellular growth by cycling between active GTP- and inactive GDP bound states. While RAS activity is modulated by factors (guanine nucleotide exchange and GTPase activating proteins) that control levels of active Ras-GTP, RAS proteins also undergo a number of post-translational modifications that regulate their function. One such modification is ubiquitylation. Monoubiquitylation of KRAS at lysine 147 (mUbRAS) enhances Ras activation and promotes signaling through the RAF and Phosphoinositide 3-Kinase (PI3K) signaling pathways. We have previously shown that mUbRAS leads to activation of RAS through a defect in GTPase activating protein (GAP) mediated downregulation, similar to the action of most oncogenic mutations. Consistent with these findings, we now show that mUbRASimpairsRAS binding to the p120 GAP catalytic domain. Mutations in activated G12V RAS that prevent ubiquitylaton at 147 show a decrease in tumorigenesis, suggesting that in addition to activating KRAS, monoubiquitylation at this site may promote downstream signaling and transformation. To investigate whether mUbRAS alters RAS effector interactions, we chemically ubiquitylated KRAS at residue 147 and characterized binding of mUbRAS to RAS binding domains (RBDs) from three distinct downstream effectors that play key roles in RAS-mediated transformation. Results from these studies show a decrease in binding of mUbRAS (7-10-fold) relative to the CRAF RAS Binding Domain (RBD), the catalytic subunit of Phosphoinositide 3-Kinase catalytic gamma (PI3Kcγ) and RALGDS RBD. Intriguingly, we find that mUbRAS shows greatly enhanced (> 40-fold) binding to the CRAF RBD when bound to GDP. These findings, taken together, suggest that mUbRASmay promoteactivation of RAS through a GAP defect, and facilitate RAF association and MAPK signaling in a nucleotide independent manner.
Keywords: RAS GTPase; allostery; effector; signal transduction; ubiquitylation.
Figures
Similar articles
-
A KRAS GTPase K104Q Mutant Retains Downstream Signaling by Offsetting Defects in Regulation.J Biol Chem. 2017 Mar 17;292(11):4446-4456. doi: 10.1074/jbc.M116.762435. Epub 2017 Jan 30. J Biol Chem. 2017. PMID: 28154176 Free PMC article.
-
Raf-1 is involved in the regulation of the interaction between guanine nucleotide exchange factor and Ha-ras. Evidences for a function of Raf-1 and phosphatidylinositol 3-kinase upstream to Ras.J Biol Chem. 1998 Dec 25;273(52):34737-44. doi: 10.1074/jbc.273.52.34737. J Biol Chem. 1998. PMID: 9856997
-
A new function of p120-GTPase-activating protein. Prevention of the guanine nucleotide exchange factor-stimulated nucleotide exchange on the active form of Ha-ras p21.J Biol Chem. 1997 Oct 3;272(40):25128-34. doi: 10.1074/jbc.272.40.25128. J Biol Chem. 1997. PMID: 9312123
-
Ras Conformational Ensembles, Allostery, and Signaling.Chem Rev. 2016 Jun 8;116(11):6607-65. doi: 10.1021/acs.chemrev.5b00542. Epub 2016 Jan 27. Chem Rev. 2016. PMID: 26815308 Review.
-
Regulation of phosphorylation pathways by p21 GTPases. The p21 Ras-related Rho subfamily and its role in phosphorylation signalling pathways.Eur J Biochem. 1996 Dec 1;242(2):171-85. doi: 10.1111/j.1432-1033.1996.0171r.x. Eur J Biochem. 1996. PMID: 8973630 Review.
Cited by
-
Monoubiquitination of KRAS at Lysine104 and Lysine147 Modulates Its Dynamics and Interaction with Partner Proteins.J Phys Chem B. 2021 May 13;125(18):4681-4691. doi: 10.1021/acs.jpcb.1c01062. Epub 2021 Apr 30. J Phys Chem B. 2021. PMID: 33929846 Free PMC article.
-
A comprehensive analysis of RAS-effector interactions reveals interaction hotspots and new binding partners.J Biol Chem. 2021 Jan-Jun;296:100626. doi: 10.1016/j.jbc.2021.100626. Epub 2021 Apr 28. J Biol Chem. 2021. PMID: 33930461 Free PMC article.
-
Functional and structural insights into RAS effector proteins.Mol Cell. 2024 Aug 8;84(15):2807-2821. doi: 10.1016/j.molcel.2024.06.027. Epub 2024 Jul 17. Mol Cell. 2024. PMID: 39025071 Free PMC article. Review.
-
Divergent Mechanisms Activating RAS and Small GTPases Through Post-translational Modification.Front Mol Biosci. 2021 Jul 8;8:707439. doi: 10.3389/fmolb.2021.707439. eCollection 2021. Front Mol Biosci. 2021. PMID: 34307463 Free PMC article. Review.
-
USP25 maintains KRAS expression and inhibiting the deubiquitinase suppresses KRAS signaling in human cancer.J Biol Chem. 2025 Jul;301(7):110337. doi: 10.1016/j.jbc.2025.110337. Epub 2025 Jun 3. J Biol Chem. 2025. PMID: 40473213 Free PMC article.
References
-
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer Statistics, 2009. CA Cancer J. Clin. 2009;59(4):225-249. https://doi.org/10.3322/caac.20006 - DOI - PubMed
-
- Hobbs GA, Der CJ, Rossman KL. RAS Isoforms and Mutations in Cancer at a Glance. J. Cell Sci. 2016;129:1287-1292. https://doi.org/10.1242/jcs.182873 - DOI - PMC - PubMed
-
- Bos JL, Rehmann H, Wittinghofer A. Review GEFs and GAPs : Critical Elements in the Control of Small G Proteins. Cell. 2007;129(5):865-877. https://doi.org/10.1016/j.cell.2007.05.018 - DOI - PubMed
-
- Du X, Sprang SR. Transition State Structures and the Roles of Catalytic Residues in GAP-Facilitated GTPase of Ras as Elucidated by (18)O Kinetic Isotope Effects. Biochemistry 2009;48(21):4538-4547. https://doi.org/10.1021/bi802359b - DOI - PMC - PubMed
-
- Scheffzek K, Ahmadian MR, Wittinghofer A. GTPase-Activating Proteins: Helping Hands to Complement an Active Site. Trends Biochem. Sci. 1998;23(7):257-262. https://doi.org/10.1016/S0968-0004(98)01224-9 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
