NuMA recruits dynein activity to microtubule minus-ends at mitosis
- PMID: 29185983
- PMCID: PMC5706958
- DOI: 10.7554/eLife.29328
NuMA recruits dynein activity to microtubule minus-ends at mitosis
Abstract
To build the spindle at mitosis, motors exert spatially regulated forces on microtubules. We know that dynein pulls on mammalian spindle microtubule minus-ends, and this localized activity at ends is predicted to allow dynein to cluster microtubules into poles. How dynein becomes enriched at minus-ends is not known. Here, we use quantitative imaging and laser ablation to show that NuMA targets dynactin to minus-ends, localizing dynein activity there. NuMA is recruited to new minus-ends independently of dynein and more quickly than dynactin; both NuMA and dynactin display specific, steady-state binding at minus-ends. NuMA localization to minus-ends involves a C-terminal region outside NuMA's canonical microtubule-binding domain and is independent of minus-end binders γ-TuRC, CAMSAP1, and KANSL1/3. Both NuMA's minus-end-binding and dynein-dynactin-binding modules are required to rescue focused, bipolar spindle organization. Thus, NuMA may serve as a mitosis-specific minus-end cargo adaptor, targeting dynein activity to minus-ends to cluster spindle microtubules into poles.
Keywords: Potorous tridactylus; cell biology; dynein; human; microtubule; minus-end; mitosis; spindle.
Conflict of interest statement
No competing interests declared.
Figures

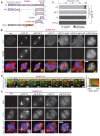
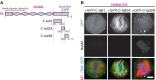
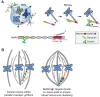

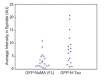
Similar articles
-
Role of NuMA in vertebrate cells: review of an intriguing multifunctional protein.Front Biosci. 2006 Jan 1;11:1137-46. doi: 10.2741/1868. Front Biosci. 2006. PMID: 16146802 Review.
-
NuMA is a mitotic adaptor protein that activates dynein and connects it to microtubule minus ends.J Cell Biol. 2025 Apr 7;224(4):e202408118. doi: 10.1083/jcb.202408118. Epub 2025 Feb 11. J Cell Biol. 2025. PMID: 39932518 Free PMC article.
-
Dynein-Dynactin-NuMA clusters generate cortical spindle-pulling forces as a multi-arm ensemble.Elife. 2018 May 31;7:e36559. doi: 10.7554/eLife.36559. Elife. 2018. PMID: 29848445 Free PMC article.
-
Force on spindle microtubule minus ends moves chromosomes.J Cell Biol. 2014 Jul 21;206(2):245-56. doi: 10.1083/jcb.201401091. Epub 2014 Jul 14. J Cell Biol. 2014. PMID: 25023517 Free PMC article.
-
NuMA: a nuclear protein involved in mitotic centrosome function.Microsc Res Tech. 2000 Jun 1;49(5):467-77. doi: 10.1002/(SICI)1097-0029(20000601)49:5<467::AID-JEMT9>3.0.CO;2-V. Microsc Res Tech. 2000. PMID: 10842374 Review.
Cited by
-
Microtubule End-Clustering Maintains a Steady-State Spindle Shape.Curr Biol. 2019 Feb 18;29(4):700-708.e5. doi: 10.1016/j.cub.2019.01.016. Epub 2019 Feb 7. Curr Biol. 2019. PMID: 30744975 Free PMC article.
-
CAMSAP2 is required for bridging fiber assembly to ensure mitotic spindle assembly and chromosome segregation in human epithelial Caco-2 cells.PLoS One. 2025 Jan 9;20(1):e0308150. doi: 10.1371/journal.pone.0308150. eCollection 2025. PLoS One. 2025. PMID: 39787108 Free PMC article.
-
The cytoplasmic dynein transport machinery and its many cargoes.Nat Rev Mol Cell Biol. 2018 Jun;19(6):382-398. doi: 10.1038/s41580-018-0004-3. Nat Rev Mol Cell Biol. 2018. PMID: 29662141 Free PMC article. Review.
-
Mammalian kinetochores count attached microtubules in a sensitive and switch-like manner.J Cell Biol. 2019 Nov 4;218(11):3583-3596. doi: 10.1083/jcb.201902105. Epub 2019 Sep 6. J Cell Biol. 2019. PMID: 31492713 Free PMC article.
-
Multiple motors cooperate to establish and maintain acentrosomal spindle bipolarity in C. elegans oocyte meiosis.Elife. 2022 Feb 11;11:e72872. doi: 10.7554/eLife.72872. Elife. 2022. PMID: 35147496 Free PMC article.
References
-
- Chinen T, Liu P, Shioda S, Pagel J, Cerikan B, Lin TC, Gruss O, Hayashi Y, Takeno H, Shima T, Okada Y, Hayakawa I, Hayashi Y, Kigoshi H, Usui T, Schiebel E. The γ-tubulin-specific inhibitor gatastatin reveals temporal requirements of microtubule nucleation during the cell cycle. Nature Communications. 2015;6:8722. doi: 10.1038/ncomms9722. - DOI - PMC - PubMed
-
- Compton DA, Luo C. Mutation of the predicted p34cdc2 phosphorylation sites in NuMA impair the assembly of the mitotic spindle and block mitosis. Journal of Cell Science. 1995;108:621–633. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

