Immune evasion of Plasmodium falciparum by RIFIN via inhibitory receptors
- PMID: 29186116
- PMCID: PMC5748893
- DOI: 10.1038/nature24994
Immune evasion of Plasmodium falciparum by RIFIN via inhibitory receptors
Erratum in
-
Corrigendum: Immune evasion of Plasmodium falciparum by RIFIN via inhibitory receptors.Nature. 2018 Feb 22;554(7693):554. doi: 10.1038/nature25498. Epub 2018 Feb 14. Nature. 2018. PMID: 29443969
Abstract
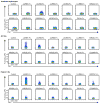
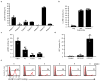
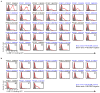
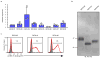
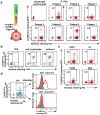
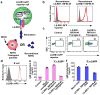
Malaria is among the most serious infectious diseases affecting humans, accounting for approximately half a million deaths each year. Plasmodium falciparum causes most life-threatening cases of malaria. Acquired immunity to malaria is inefficient, even after repeated exposure to P. falciparum, but the immune regulatory mechanisms used by P. falciparum remain largely unknown. Here we show that P. falciparum uses immune inhibitory receptors to achieve immune evasion. RIFIN proteins are products of a polymorphic multigene family comprising approximately 150-200 genes per parasite genome that are expressed on the surface of infected erythrocytes. We found that a subset of RIFINs binds to either leucocyte immunoglobulin-like receptor B1 (LILRB1) or leucocyte-associated immunoglobulin-like receptor 1 (LAIR1). LILRB1-binding RIFINs inhibit activation of LILRB1-expressing B cells and natural killer (NK) cells. Furthermore, P. falciparum-infected erythrocytes isolated from patients with severe malaria were more likely to interact with LILRB1 than erythrocytes from patients with non-severe malaria, although an extended study with larger sample sizes is required to confirm this finding. Our results suggest that P. falciparum has acquired multiple RIFINs to evade the host immune system by targeting immune inhibitory receptors.
Figures

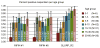


References
-
- World Health Organization. World malaria report 2016. World Health Organization; 2016.
-
- Langhorne J, Ndungu FM, Sponaas AM, Marsh K. Immunity to malaria: more questions than answers. Nat Immonol. 2008;9:725–732. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

