A putative R3 MYB repressor is the candidate gene underlying atroviolacium, a locus for anthocyanin pigmentation in tomato fruit
- PMID: 29186488
- PMCID: PMC5854135
- DOI: 10.1093/jxb/erx382
A putative R3 MYB repressor is the candidate gene underlying atroviolacium, a locus for anthocyanin pigmentation in tomato fruit
Abstract
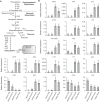
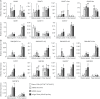
Anthocyanins are potential health-promoting compounds in the human diet. The atv (atroviolacium) locus, derived from the wild tomato species Solanum cheesmaniae, has been shown to enhance anthocyanin pigmentation in tomato fruit when it co-exists with either the Aft (Anthocyanin fruit) or the Abg (Aubergine) locus. In the present study, the atv locus was fine-mapped to an approximately 5.0-kb interval on chromosome 7. A putative R3 MYB repressor was identified in this interval and is hereby designated as SlMYBATV. The allele of SlMYBATV underlying the atv locus harbored a 4-bp insertion in its coding region, which is predicted to result in a frame-shift and premature protein truncation. The other candidate R3 MYB and R2R3 MYB repressors of anthocyanin biosynthesis were also identified in tomato via a genome-wide search. Transcriptional analysis showed that most of the structural genes and several regulatory genes of anthocyanin biosynthesis were up-regulated in the tomato SlMYBATV mutant lines. These findings may facilitate the elucidation of the molecular mechanisms underlying anthocyanin pigmentation in tomato fruit and help in the marker-assisted selection of anthocyanin-enriched tomato cultivars.
Keywords: Anthocyanin; MYB repressor; SlMYBATV; atroviolacium; fine-mapping; tomato; transcriptional analysis.
© The Author 2017. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures





References
-
- Aharoni A, De Vos CH, Wein M, Sun Z, Greco R, Kroon A, Mol JN, O’Connell AP. 2001. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. The Plant Journal 28, 319–332. - PubMed
-
- Al-Sane KO, Povero G, Perata P. 2011. Anthocyanin tomato mutants: overview and characterization of an anthocyanin-less somaclonal mutant. Plant Biosystems 145, 436–444.
-
- Albert NW, Lewis DH, Zhang H, Schwinn KE, Jameson PE, Davies KM. 2011. Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. The Plant Journal 65, 771–784. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

