The Planteome database: an integrated resource for reference ontologies, plant genomics and phenomics
- PMID: 29186578
- PMCID: PMC5753347
- DOI: 10.1093/nar/gkx1152
The Planteome database: an integrated resource for reference ontologies, plant genomics and phenomics
Abstract
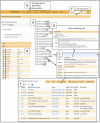
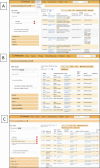
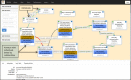
The Planteome project (http://www.planteome.org) provides a suite of reference and species-specific ontologies for plants and annotations to genes and phenotypes. Ontologies serve as common standards for semantic integration of a large and growing corpus of plant genomics, phenomics and genetics data. The reference ontologies include the Plant Ontology, Plant Trait Ontology and the Plant Experimental Conditions Ontology developed by the Planteome project, along with the Gene Ontology, Chemical Entities of Biological Interest, Phenotype and Attribute Ontology, and others. The project also provides access to species-specific Crop Ontologies developed by various plant breeding and research communities from around the world. We provide integrated data on plant traits, phenotypes, and gene function and expression from 95 plant taxa, annotated with reference ontology terms. The Planteome project is developing a plant gene annotation platform; Planteome Noctua, to facilitate community engagement. All the Planteome ontologies are publicly available and are maintained at the Planteome GitHub site (https://github.com/Planteome) for sharing, tracking revisions and new requests. The annotated data are freely accessible from the ontology browser (http://browser.planteome.org/amigo) and our data repository.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Avraham S., Tung C.-W., Ilic K., Jaiswal P., Kellogg E.A., McCouch S., Pujar A., Reiser L., Rhee S.Y., Sachs M.M. et al. . The Plant Ontology Database: a community resource for plant structure and developmental stages controlled vocabulary and annotations. Nucleic Acids Res. 2008; 36:D449–D454. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

