Type-B ARRs Control Carpel Regeneration Through Mediating AGAMOUS Expression in Arabidopsis
- PMID: 29186581
- PMCID: PMC6018948
- DOI: 10.1093/pcp/pcx187
Type-B ARRs Control Carpel Regeneration Through Mediating AGAMOUS Expression in Arabidopsis
Erratum in
-
Erratum.Plant Cell Physiol. 2018 Apr 1;59(4):876. doi: 10.1093/pcp/pcy075. Plant Cell Physiol. 2018. PMID: 29718476 Free PMC article. No abstract available.
Abstract
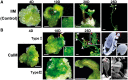
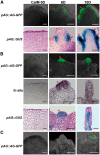
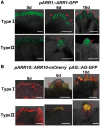
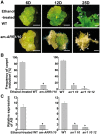
Plants are known for their capacity to regenerate organs, such as shoot, root and floral organs. Recently, a number of studies contributed to understanding the mechanisms of shoot and root regeneration. However, the mechanisms underlying floral organ regeneration are largely unknown. In this study, we established a carpel regeneration system in which two types of carpels were induced by exogenous cytokinin. For type I, all the floral organs in the regenerated inflorescence were transformed into carpels. For type II, carpels were generated directly from callus. The transcript level of AGAMOUS (AG), the carpel identity gene, was up-regulated during carpel induction. The expression signals of AG were detected in the initiating carpel primordia and regenerating carpels, and co-localized with those of two Type-B ARABIDOPSIS RESPONSE REGULATORs (ARRs), ARR1 and ARR10. Repression of either AG or type-B ARRs reduced carpel regeneration. Binding analyses showed that ARR1 and ARR10 directly bound to transcriptional regulatory regions of AG and positively regulated its expression. In addition, the expression of type-B ARRs overlapped with that of AG in the floral primordia in planta. Defects in type-B ARRs reduced the number of carpels. The results indicate that type-B ARRs control carpel regeneration through activating AG expression. Our results provide new information for understanding the mechanism of carpel formation.
Figures



References
-
- An Y.R., Li X.G., Su H.Y., Zhang X.S. (2004) Pistil induction by hormones from the callus of Oryza sativa in vitro. Plant Cell Rep. 23: 448–452. - PubMed
-
- Bertrand C., Bergounioux C., Domenichini S., Delarue M., Zhou D.X. (2003) Arabidopsis histone acetyltransferase AtGCN5 regulates the floral meristem activity through the WUSCHEL/AGAMOUS pathway. J. Biol. Chem. 278: 28246–2851. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

