Aurora A kinase phosphorylates Hec1 to regulate metaphase kinetochore-microtubule dynamics
- PMID: 29187526
- PMCID: PMC5748988
- DOI: 10.1083/jcb.201707160
Aurora A kinase phosphorylates Hec1 to regulate metaphase kinetochore-microtubule dynamics
Abstract
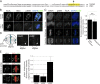
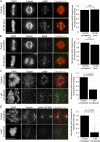
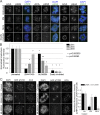
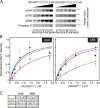
Precise regulation of kinetochore-microtubule attachments is essential for successful chromosome segregation. Central to this regulation is Aurora B kinase, which phosphorylates kinetochore substrates to promote microtubule turnover. A critical target of Aurora B is the N-terminal "tail" domain of Hec1, which is a component of the NDC80 complex, a force-transducing link between kinetochores and microtubules. Although Aurora B is regarded as the "master regulator" of kinetochore-microtubule attachment, other mitotic kinases likely contribute to Hec1 phosphorylation. In this study, we demonstrate that Aurora A kinase regulates kinetochore-microtubule dynamics of metaphase chromosomes, and we identify Hec1 S69, a previously uncharacterized phosphorylation target site in the Hec1 tail, as a critical Aurora A substrate for this regulation. Additionally, we demonstrate that Aurora A kinase associates with inner centromere protein (INCENP) during mitosis and that INCENP is competent to drive accumulation of the kinase to the centromere region of mitotic chromosomes. These findings reveal that both Aurora A and B contribute to kinetochore-microtubule attachment dynamics, and they uncover an unexpected role for Aurora A in late mitosis.
© 2018 DeLuca et al.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

