Epitope-focused immunogens against the CD4-binding site of HIV-1 envelope protein induce neutralizing antibodies against auto- and heterologous viruses
- PMID: 29187598
- PMCID: PMC5777257
- DOI: 10.1074/jbc.M117.816447
Epitope-focused immunogens against the CD4-binding site of HIV-1 envelope protein induce neutralizing antibodies against auto- and heterologous viruses
Abstract
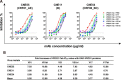
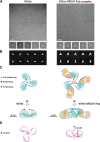
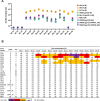
Recent discoveries of broadly neutralizing antibodies (bnAbs) in HIV-1-infected individuals have led to the identification of several major "vulnerable sites" on the HIV-1 envelope (Env) glycoprotein. These sites have provided precise targets for HIV-1 vaccine development, but identifying and utilizing many of these targets remain technically challenging. Using a yeast surface display-based approach, we sought to identify epitope-focused antigenic domains (EADs) containing one of the "vulnerable sites," the CD4-binding site (CD4bs), through screening and selection of a combinatorial antigen library of the HIV-1 envelope glycoprotein with the CD4bs bnAb VRC01. We isolated multiple EADs and found that their trimeric forms have biochemical and structural features that preferentially bind and activate B cells that express VRC01 in vitro More importantly, these EADs could induce detectable levels of neutralizing antibodies against genetically related autologous and heterologous subtype B viruses in guinea pigs. Our results demonstrate that an epitope-focused approach involving a screen of a combinatorial antigen library is feasible. The EADs identified here represent a promising collection of possible targets in the rational design of HIV-1 vaccines and lay the foundation for harnessing the specific antigenicity of CD4bs for protective immunogenicity in vivo.
Keywords: CD4bs; V3; antibody; electron microscopy (EM); epitope; guinea pig; human immunodeficiency virus (HIV); vaccine; yeast.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

