Single-cut genome editing restores dystrophin expression in a new mouse model of muscular dystrophy
- PMID: 29187645
- PMCID: PMC5749406
- DOI: 10.1126/scitranslmed.aan8081
Single-cut genome editing restores dystrophin expression in a new mouse model of muscular dystrophy
Erratum in
-
Erratum for the Research Article: "Single-cut genome editing restores dystrophin expression in a new mouse model of muscular dystrophy" by L. Amoasii, C. Long, H. Li, A. A. Mireault, J. M. Shelton, E. Sanchez-Ortiz, J. R. McAnally, S. Bhattacharyya, F. Schmidt, D. Grimm, S. D. Hauschka, R. Bassel-Duby, E. N. Olson.Sci Transl Med. 2018 Jan 24;10(425):eaat0240. doi: 10.1126/scitranslmed.aat0240. Sci Transl Med. 2018. PMID: 29367344 No abstract available.
Abstract
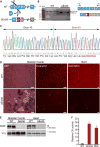
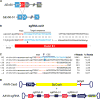
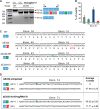
Duchenne muscular dystrophy (DMD) is a severe, progressive muscle disease caused by mutations in the dystrophin gene. The majority of DMD mutations are deletions that prematurely terminate the dystrophin protein. Deletions of exon 50 of the dystrophin gene are among the most common single exon deletions causing DMD. Such mutations can be corrected by skipping exon 51, thereby restoring the dystrophin reading frame. Using clustered regularly interspaced short palindromic repeats/CRISPR-associated 9 (CRISPR/Cas9), we generated a DMD mouse model by deleting exon 50. These ΔEx50 mice displayed severe muscle dysfunction, which was corrected by systemic delivery of adeno-associated virus encoding CRISPR/Cas9 genome editing components. We optimized the method for dystrophin reading frame correction using a single guide RNA that created reframing mutations and allowed skipping of exon 51. In conjunction with muscle-specific expression of Cas9, this approach restored up to 90% of dystrophin protein expression throughout skeletal muscles and the heart of ΔEx50 mice. This method of permanently bypassing DMD mutations using a single cut in genomic DNA represents a step toward clinical correction of DMD mutations and potentially those of other neuromuscular disorders.
Copyright © 2017 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures




References
-
- O’Brien KF, Kunkel LM. Dystrophin and muscular dystrophy: Past, present, and future. Mol Gen Metab. 2001;74:75–88. - PubMed
-
- Muntoni F, Torelli S, Ferlini A. Dystrophin and mutations: One gene, several proteins, multiple phenotypes. Lancet Neurol. 2003;2:731–740. - PubMed
-
- Mercuri E, Muntoni F. Muscular dystrophies. Lancet. 2013;381:845–860. - PubMed
-
- Campbell KP, Kahl SD. Association of dystrophin and an integral membrane glycoprotein. Nature. 1989;338:259–262. - PubMed
-
- Ervasti JM, Ohlendieck K, Kahl SD, Gaver MG, Campbell KP. Deficiency of a glycoprotein component of the dystrophin complex in dystrophic muscle. Nature. 1990;345:315–319. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

