Genome Comparison of Erythromycin Resistant Campylobacter from Turkeys Identifies Hosts and Pathways for Horizontal Spread of erm(B) Genes
- PMID: 29187841
- PMCID: PMC5695097
- DOI: 10.3389/fmicb.2017.02240
Genome Comparison of Erythromycin Resistant Campylobacter from Turkeys Identifies Hosts and Pathways for Horizontal Spread of erm(B) Genes
Abstract
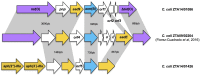
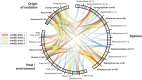
Pathogens in the genus Campylobacter are the most common cause of food-borne bacterial gastro-enteritis. Campylobacteriosis, caused principally by Campylobacter jejuni and Campylobacter coli, is transmitted to humans by food of animal origin, especially poultry. As for many pathogens, antimicrobial resistance in Campylobacter is increasing at an alarming rate. Erythromycin prescription is the treatment of choice for clinical cases requiring antimicrobial therapy but this is compromised by mobility of the erythromycin resistance gene erm(B) between strains. Here, we evaluate resistance to six antimicrobials in 170 Campylobacter isolates (133 C. coli and 37 C. jejuni) from turkeys. Erythromycin resistant isolates (n = 85; 81 C. coli and 4 C. jejuni) were screened for the presence of the erm(B) gene, that has not previously been identified in isolates from turkeys. The genomes of two positive C. coli isolates were sequenced and in both isolates the erm(B) gene clustered with resistance determinants against aminoglycosides plus tetracycline, including aad9, aadE, aph(2″)-IIIa, aph(3')-IIIa, and tet(O) genes. Comparative genomic analysis identified identical erm(B) sequences among Campylobacter from turkeys, Streptococcus suis from pigs and Enterococcus faecium and Clostridium difficile from humans. This is consistent with multiple horizontal transfer events among different bacterial species colonizing turkeys. This example highlights the potential for dissemination of antimicrobial resistance across bacterial species boundaries which may compromise their effectiveness in antimicrobial therapy.
Keywords: Campylobacter; antimicrobial; erm(B); erythromycin; transmission; turkey.
Figures
References
-
- Chapman J. S. (2003). Disinfectant resistance mechanisms, cross-resistance, and co-resistance. Int. Biodeterior. Biodegradation 51 271–276. 10.1016/S0964-8305(03)00044-1 - DOI
-
- Chen J., Yu Z., Michel F. C., Jr., Wittum T., Morrison M. (2007). Development and application of real-time PCR assays for quantification of erm genes conferring resistance to macrolides-lincosamides-streptogramin B in livestock manure and manure management systems. Appl. Environ. Microbiol. 73 4407–4416. 10.1128/AEM.02799-06 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

