A semi-synthetic organism that stores and retrieves increased genetic information
- PMID: 29189780
- PMCID: PMC5796663
- DOI: 10.1038/nature24659
A semi-synthetic organism that stores and retrieves increased genetic information
Abstract
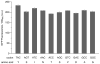
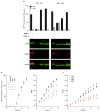
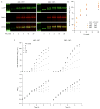
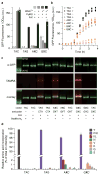
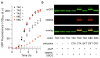
Since at least the last common ancestor of all life on Earth, genetic information has been stored in a four-letter alphabet that is propagated and retrieved by the formation of two base pairs. The central goal of synthetic biology is to create new life forms and functions, and the most general route to this goal is the creation of semi-synthetic organisms whose DNA harbours two additional letters that form a third, unnatural base pair. Previous efforts to generate such semi-synthetic organisms culminated in the creation of a strain of Escherichia coli that, by virtue of a nucleoside triphosphate transporter from Phaeodactylum tricornutum, imports the requisite unnatural triphosphates from its medium and then uses them to replicate a plasmid containing the unnatural base pair dNaM-dTPT3. Although the semi-synthetic organism stores increased information when compared to natural organisms, retrieval of the information requires in vivo transcription of the unnatural base pair into mRNA and tRNA, aminoacylation of the tRNA with a non-canonical amino acid, and efficient participation of the unnatural base pair in decoding at the ribosome. Here we report the in vivo transcription of DNA containing dNaM and dTPT3 into mRNAs with two different unnatural codons and tRNAs with cognate unnatural anticodons, and their efficient decoding at the ribosome to direct the site-specific incorporation of natural or non-canonical amino acids into superfolder green fluorescent protein. The results demonstrate that interactions other than hydrogen bonding can contribute to every step of information storage and retrieval. The resulting semi-synthetic organism both encodes and retrieves increased information and should serve as a platform for the creation of new life forms and functions.
Conflict of interest statement
The authors declare the following competing financial interests: A provisional patent application has been filed by Synthorx and The Scripps Research Institute (application #62/531,325; inventors J.L.P, C.E.C., H.R.A, Y.Z., E.C.F., A.W.F., V.T. Dien, and F.E.R.) covering the use of UBPs in tRNAs and mRNAs to produce proteins containing ncAAs. J.L.P, C.E.C., H.R.A., K.S.J., and F.E.R. have a financial interest (shares) in Synthorx, Inc., a company that has commercial interests in the UBP.
Figures
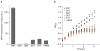



Comment in
-
'Alien' DNA makes proteins in living cells for the first time.Nature. 2017 Nov 29;551(7682):550-551. doi: 10.1038/nature.2017.23040. Nature. 2017. PMID: 29189787 No abstract available.
Similar articles
-
Progress Toward a Semi-Synthetic Organism with an Unrestricted Expanded Genetic Alphabet.J Am Chem Soc. 2018 Nov 28;140(47):16115-16123. doi: 10.1021/jacs.8b08416. Epub 2018 Nov 12. J Am Chem Soc. 2018. PMID: 30418780 Free PMC article.
-
New codons for efficient production of unnatural proteins in a semisynthetic organism.Nat Chem Biol. 2020 May;16(5):570-576. doi: 10.1038/s41589-020-0507-z. Epub 2020 Apr 6. Nat Chem Biol. 2020. PMID: 32251411 Free PMC article.
-
A semi-synthetic organism with an expanded genetic alphabet.Nature. 2014 May 15;509(7500):385-8. doi: 10.1038/nature13314. Epub 2014 May 7. Nature. 2014. PMID: 24805238 Free PMC article.
-
The expanded genetic alphabet.Angew Chem Int Ed Engl. 2015 Oct 5;54(41):11930-44. doi: 10.1002/anie.201502890. Epub 2015 Aug 25. Angew Chem Int Ed Engl. 2015. PMID: 26304162 Free PMC article. Review.
-
Creation, Optimization, and Use of Semi-Synthetic Organisms that Store and Retrieve Increased Genetic Information.J Mol Biol. 2022 Apr 30;434(8):167331. doi: 10.1016/j.jmb.2021.167331. Epub 2021 Oct 25. J Mol Biol. 2022. PMID: 34710400 Review.
Cited by
-
Alternative Biochemistries for Alien Life: Basic Concepts and Requirements for the Design of a Robust Biocontainment System in Genetic Isolation.Genes (Basel). 2018 Dec 28;10(1):17. doi: 10.3390/genes10010017. Genes (Basel). 2018. PMID: 30597824 Free PMC article. Review.
-
DNA-based memory devices for recording cellular events.Nat Rev Genet. 2018 Nov;19(11):718-732. doi: 10.1038/s41576-018-0052-8. Nat Rev Genet. 2018. PMID: 30237447 Free PMC article. Review.
-
The Role of Orthogonality in Genetic Code Expansion.Life (Basel). 2019 Jul 5;9(3):58. doi: 10.3390/life9030058. Life (Basel). 2019. PMID: 31284384 Free PMC article.
-
Engineering tRNAs for the Ribosomal Translation of Non-proteinogenic Monomers.Chem Rev. 2024 May 22;124(10):6444-6500. doi: 10.1021/acs.chemrev.3c00894. Epub 2024 Apr 30. Chem Rev. 2024. PMID: 38688034 Free PMC article. Review.
-
Genetic Alphabet Expansion Provides Versatile Specificities and Activities of Unnatural-Base DNA Aptamers Targeting Cancer Cells.Mol Ther Nucleic Acids. 2019 Mar 1;14:158-170. doi: 10.1016/j.omtn.2018.11.011. Epub 2018 Nov 29. Mol Ther Nucleic Acids. 2019. PMID: 30594072 Free PMC article.
References
-
- Leduc S. The Mechanisms of Life. Rebman Company; 1911.
-
- Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24:79–88. - PubMed
-
- Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–444. - PubMed

