Transcriptome profiling analysis of senescent gingival fibroblasts in response to Fusobacterium nucleatum infection
- PMID: 29190775
- PMCID: PMC5708803
- DOI: 10.1371/journal.pone.0188755
Transcriptome profiling analysis of senescent gingival fibroblasts in response to Fusobacterium nucleatum infection
Erratum in
-
Correction: Transcriptome profiling analysis of senescent gingival fibroblasts in response to Fusobacterium nucleatum infection.PLoS One. 2018 Jan 9;13(1):e0191209. doi: 10.1371/journal.pone.0191209. eCollection 2018. PLoS One. 2018. PMID: 29315353 Free PMC article.
Abstract
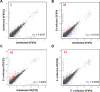
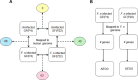
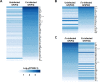
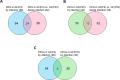
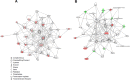
Periodontal disease is caused by dental plaque biofilms. Fusobacterium nucleatum is an important periodontal pathogen involved in the development of bacterial complexity in dental plaque biofilms. Human gingival fibroblasts (GFs) act as the first line of defense against oral microorganisms and locally orchestrate immune responses by triggering the production of reactive oxygen species and pro-inflammatory cytokines (IL-6 and IL-8). The frequency and severity of periodontal diseases is known to increase in elderly subjects. However, despite several studies exploring the effects of aging in periodontal disease, the underlying mechanisms through which aging affects the interaction between F. nucleatum and human GFs remain unclear. To identify genes affected by infection, aging, or both, we performed an RNA-Seq analysis using GFs isolated from a single healthy donor that were passaged for a short period of time (P4) 'young GFs' or for longer period of time (P22) 'old GFs', and infected or not with F. nucleatum. Comparing F. nucleatum-infected and uninfected GF(P4) cells the differentially expressed genes (DEGs) were involved in host defense mechanisms (i.e., immune responses and defense responses), whereas comparing F. nucleatum-infected and uninfected GF(P22) cells the DEGs were involved in cell maintenance (i.e., TGF-β signaling, skeletal development). Most DEGs in F. nucleatum-infected GF(P22) cells were downregulated (85%) and were significantly associated with host defense responses such as inflammatory responses, when compared to the DEGs in F. nucleatum-infected GF(P4) cells. Five genes (GADD45b, KLF10, CSRNP1, ID1, and TM4SF1) were upregulated in response to F. nucleatum infection; however, this effect was only seen in GF(P22) cells. The genes identified here appear to interact with each other in a network associated with free radical scavenging, cell cycle, and cancer; therefore, they could be potential candidates involved in the aged GF's response to F. nucleatum infection. Further studies are needed to confirm these observations.
Conflict of interest statement
Figures

References
-
- Armitage GC. Development of a classification system for periodontal diseases and conditions. Ann Periodontol. 1999;4(1):1–6. doi: 10.1902/annals.1999.4.1.1 . - DOI - PubMed
-
- Kolenbrander PE. Oral microbial communities: biofilms, interactions, and genetic systems. Annu Rev Microbiol. 2000;54:413–37. doi: 10.1146/annurev.micro.54.1.413 . - DOI - PubMed
-
- van Winkelhoff AJ, Loos BG, van der Reijden WA, van der Velden U. Porphyromonas gingivalis, Bacteroides forsythus and other putative periodontal pathogens in subjects with and without periodontal destruction. Journal of clinical periodontology. 2002;29(11):1023–8. . - PubMed
-
- Diaz PI, Zilm PS, Rogers AH. Fusobacterium nucleatum supports the growth of Porphyromonas gingivalis in oxygenated and carbon-dioxide-depleted environments. Microbiology. 2002;148(Pt 2):467–72. doi: 10.1099/00221287-148-2-467 . - DOI - PubMed
-
- Signat B, Roques C, Poulet P, Duffaut D. Fusobacterium nucleatum in periodontal health and disease. Current issues in molecular biology. 2011;13(2):25–36. . - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

