Decontaminating eukaryotic genome assemblies with machine learning
- PMID: 29191179
- PMCID: PMC5709863
- DOI: 10.1186/s12859-017-1941-0
Decontaminating eukaryotic genome assemblies with machine learning
Abstract
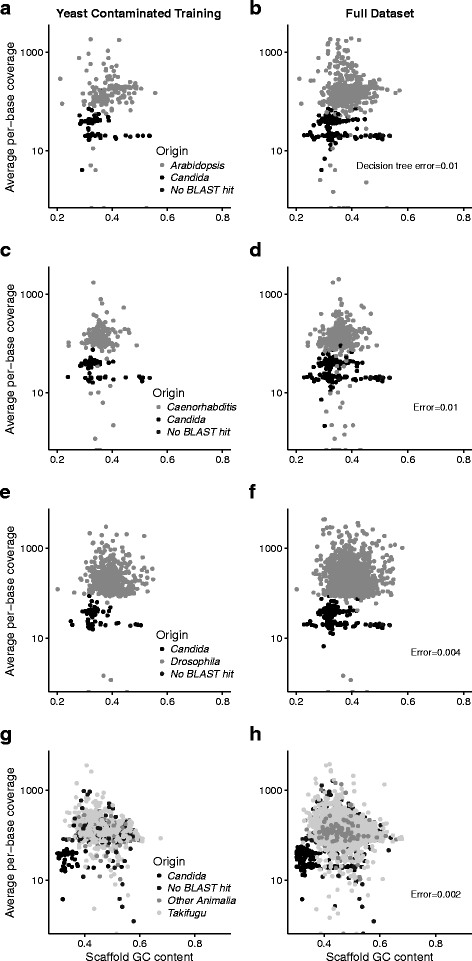
Background: High-throughput sequencing has made it theoretically possible to obtain high-quality de novo assembled genome sequences but in practice DNA extracts are often contaminated with sequences from other organisms. Currently, there are few existing methods for rigorously decontaminating eukaryotic assemblies. Those that do exist filter sequences based on nucleotide similarity to contaminants and risk eliminating sequences from the target organism.
Results: We introduce a novel application of an established machine learning method, a decision tree, that can rigorously classify sequences. The major strength of the decision tree is that it can take any measured feature as input and does not require a priori identification of significant descriptors. We use the decision tree to classify de novo assembled sequences and compare the method to published protocols.
Conclusions: A decision tree performs better than existing methods when classifying sequences in eukaryotic de novo assemblies. It is efficient, readily implemented, and accurately identifies target and contaminant sequences. Importantly, a decision tree can be used to classify sequences according to measured descriptors and has potentially many uses in distilling biological datasets.
Keywords: Contamination; DNA sequencing; Genome assembly; High-throughput; Sequence filtering.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures








References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

