Domestication of self-splicing introns during eukaryogenesis: the rise of the complex spliceosomal machinery
- PMID: 29191215
- PMCID: PMC5709842
- DOI: 10.1186/s13062-017-0201-6
Domestication of self-splicing introns during eukaryogenesis: the rise of the complex spliceosomal machinery
Abstract
ᅟ: The spliceosome is a eukaryote-specific complex that is essential for the removal of introns from pre-mRNA. It consists of five small nuclear RNAs (snRNAs) and over a hundred proteins, making it one of the most complex molecular machineries. Most of this complexity has emerged during eukaryogenesis, a period that is characterised by a drastic increase in cellular and genomic complexity. Although not fully resolved, recent findings have started to shed some light on how and why the spliceosome originated. In this paper we review how the spliceosome has evolved and discuss its origin and subsequent evolution in light of different general hypotheses on the evolution of complexity. Comparative analyses have established that the catalytic core of this ribonucleoprotein (RNP) complex, as well as the spliceosomal introns, evolved from self-splicing group II introns. Most snRNAs evolved from intron fragments and the essential Prp8 protein originated from the protein that is encoded by group II introns. Proteins that functioned in other RNA processes were added to this core and extensive duplications of these proteins substantially increased the complexity of the spliceosome prior to the eukaryotic diversification. The splicing machinery became even more complex in animals and plants, yet was simplified in eukaryotes with streamlined genomes. Apparently, the spliceosome did not evolve its complexity gradually, but in rapid bursts, followed by stagnation or even simplification. We argue that although both adaptive and neutral evolution have been involved in the evolution of the spliceosome, especially the latter was responsible for the emergence of an enormously complex eukaryotic splicing machinery from simple self-splicing sequences.
Reviewers: This article was reviewed by W. Ford Doolittle, Eugene V. Koonin and Vivek Anantharaman.
Keywords: Eukaryogenesis; Evolution of complexity; Introns; Spliceosome; Splicing.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

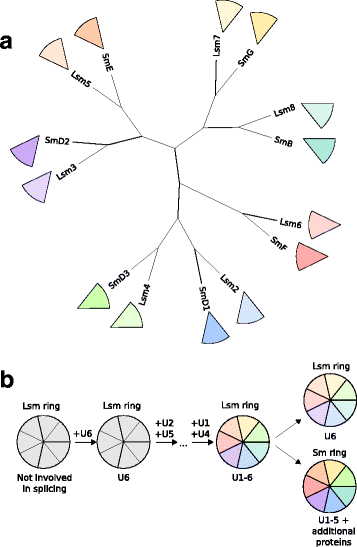
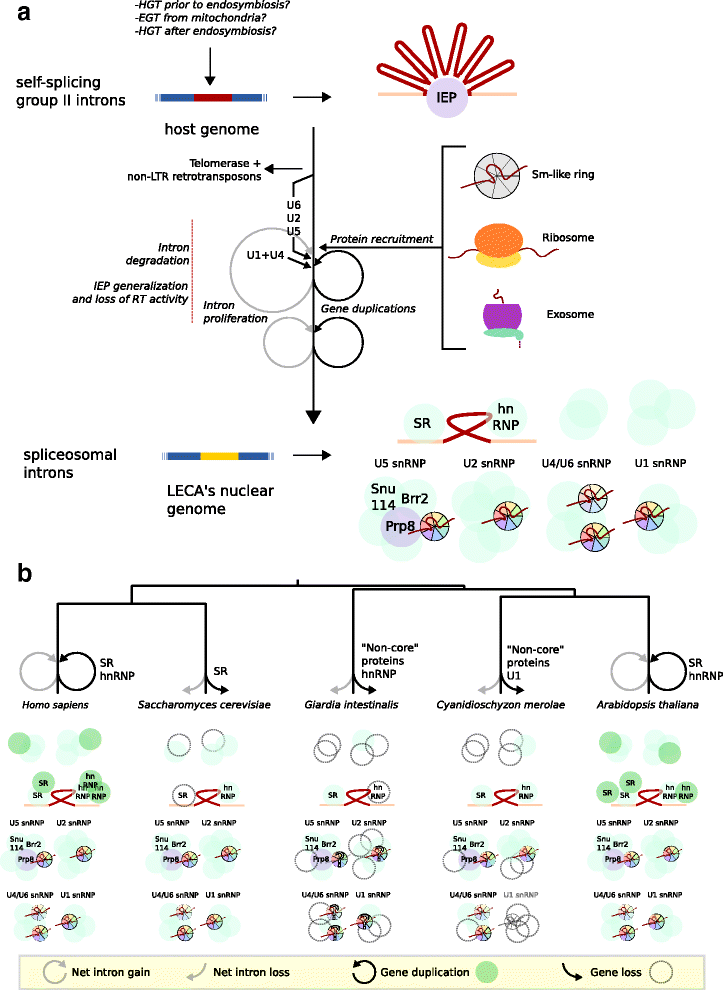
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

