Fine control of metal concentrations is necessary for cells to discern zinc from cobalt
- PMID: 29192165
- PMCID: PMC5709419
- DOI: 10.1038/s41467-017-02085-z
Fine control of metal concentrations is necessary for cells to discern zinc from cobalt
Abstract
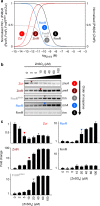
Bacteria possess transcription factors whose DNA-binding activity is altered upon binding to specific metals, but metal binding is not specific in vitro. Here we show that tight regulation of buffered intracellular metal concentrations is a prerequisite for metal specificity of Zur, ZntR, RcnR and FrmR in Salmonella Typhimurium. In cells, at non-inhibitory elevated concentrations, Zur and ZntR, only respond to Zn(II), RcnR to cobalt and FrmR to formaldehyde. However, in vitro all these sensors bind non-cognate metals, which alters DNA binding. We model the responses of these sensors to intracellular-buffered concentrations of Co(II) and Zn(II) based upon determined abundances, metal affinities and DNA affinities of each apo- and metalated sensor. The cognate sensors are modelled to respond at the lowest concentrations of their cognate metal, explaining specificity. However, other sensors are modelled to respond at concentrations only slightly higher, and cobalt or Zn(II) shock triggers mal-responses that match these predictions. Thus, perfect metal specificity is fine-tuned to a narrow range of buffered intracellular metal concentrations.
Conflict of interest statement
This work was supported by Biotechnology and Biological Research Council awards BB/J017787/1 and BB/L009226/1 to N.J.R. in conjunction with a financial contribution by Procter and Gamble (in association with BB/J017787/1 Industrial Partnership Award). The remaining authors declare no competing financial interests.
Figures

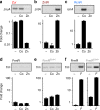
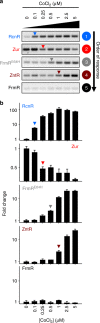
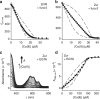
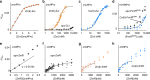



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

