Conservation of Nonsense-Mediated mRNA Decay Complex Components Throughout Eukaryotic Evolution
- PMID: 29192227
- PMCID: PMC5709506
- DOI: 10.1038/s41598-017-16942-w
Conservation of Nonsense-Mediated mRNA Decay Complex Components Throughout Eukaryotic Evolution
Abstract
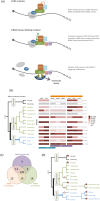
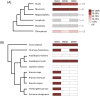
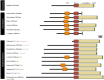
Nonsense-mediated mRNA decay (NMD) is an essential eukaryotic process regulating transcript quality and abundance, and is involved in diverse processes including brain development and plant defenses. Although some of the NMD machinery is conserved between kingdoms, little is known about its evolution. Phosphorylation of the core NMD component UPF1 is critical for NMD and is regulated in mammals by the SURF complex (UPF1, SMG1 kinase, SMG8, SMG9 and eukaryotic release factors). However, since SMG1 is reportedly missing from the genomes of fungi and the plant Arabidopsis thaliana, it remains unclear how UPF1 is activated outside the metazoa. We used comparative genomics to determine the conservation of the NMD pathway across eukaryotic evolution. We show that SURF components are present in all major eukaryotic lineages, including fungi, suggesting that in addition to UPF1 and SMG1, SMG8 and SMG9 also existed in the last eukaryotic common ancestor, 1.8 billion years ago. However, despite the ancient origins of the SURF complex, we also found that SURF factors have been independently lost across the Eukarya, pointing to genetic buffering within the essential NMD pathway. We infer an ancient role for SURF in regulating UPF1, and the intriguing possibility of undiscovered NMD regulatory pathways.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
SMG1 is an ancient nonsense-mediated mRNA decay effector.Plant J. 2013 Dec;76(5):800-10. doi: 10.1111/tpj.12329. Epub 2013 Nov 5. Plant J. 2013. PMID: 24103012
-
CK2-mediated TEL2 phosphorylation augments nonsense-mediated mRNA decay (NMD) by increase of SMG1 stability.Biochim Biophys Acta. 2013 Oct;1829(10):1047-55. doi: 10.1016/j.bbagrm.2013.06.002. Epub 2013 Jul 3. Biochim Biophys Acta. 2013. PMID: 23831331
-
Structures of SMG1-UPFs complexes: SMG1 contributes to regulate UPF2-dependent activation of UPF1 in NMD.Structure. 2014 Aug 5;22(8):1105-1119. doi: 10.1016/j.str.2014.05.015. Epub 2014 Jul 4. Structure. 2014. PMID: 25002321
-
The evolution and diversity of the nonsense-mediated mRNA decay pathway.F1000Res. 2018 Aug 15;7:1299. doi: 10.12688/f1000research.15872.2. eCollection 2018. F1000Res. 2018. PMID: 30345031 Free PMC article. Review.
-
Nonsense-mediated mRNA decay: The challenge of telling right from wrong in a complex transcriptome.Wiley Interdiscip Rev RNA. 2019 Nov;10(6):e1548. doi: 10.1002/wrna.1548. Epub 2019 May 26. Wiley Interdiscip Rev RNA. 2019. PMID: 31131562 Free PMC article. Review.
Cited by
-
Small-molecule eRF3a degraders rescue CFTR nonsense mutations by promoting premature termination codon readthrough.J Clin Invest. 2022 Sep 15;132(18):e154571. doi: 10.1172/JCI154571. J Clin Invest. 2022. PMID: 35900863 Free PMC article.
-
Restriction of an intron size en route to endothermy.Nucleic Acids Res. 2021 Mar 18;49(5):2460-2487. doi: 10.1093/nar/gkab046. Nucleic Acids Res. 2021. PMID: 33550394 Free PMC article.
-
Multi-omics analysis of SFTS virus infection in Rhipicephalus microplus cells reveals antiviral tick factors.Nat Commun. 2025 May 21;16(1):4732. doi: 10.1038/s41467-025-59565-w. Nat Commun. 2025. PMID: 40399277 Free PMC article.
-
Gene Body Methylation in Plants: Mechanisms, Functions, and Important Implications for Understanding Evolutionary Processes.Genome Biol Evol. 2022 Apr 10;14(4):evac038. doi: 10.1093/gbe/evac038. Genome Biol Evol. 2022. PMID: 35298639 Free PMC article. Review.
-
The loss of SMG1 causes defects in quality control pathways in Physcomitrella patens.Nucleic Acids Res. 2018 Jun 20;46(11):5822-5836. doi: 10.1093/nar/gky225. Nucleic Acids Res. 2018. PMID: 29596649 Free PMC article.
References
-
- Chiba Y, Green PJ. mRNA degradation machinery in plants. J. Plant Biol. 2009;52:114–124. doi: 10.1007/s12374-009-9021-2. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

