Profiling of different pancreatic cancer cells used as models for metastatic behaviour shows large variation in their N-glycosylation
- PMID: 29192278
- PMCID: PMC5709460
- DOI: 10.1038/s41598-017-16811-6
Profiling of different pancreatic cancer cells used as models for metastatic behaviour shows large variation in their N-glycosylation
Abstract
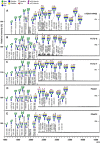
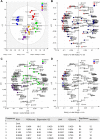
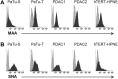
To characterise pancreatic cancer cells from different sources which are used as model systems to study the metastatic behaviour in pancreatic ductal adenocarcinoma (PDAC), we compared the N-glycan imprint of four PDAC cells which were previously shown to differ in their galectin-4 expression and metastatic potential in vivo. Next to the sister cell lines Pa-Tu-8988S and Pa-Tu-8988T, which were isolated from the same liver metastasis of a PDAC, this included two primary PDAC cell cultures, PDAC1 and PDAC2. Additionally, we extended the N-glycan profiling to a normal, immortalized pancreatic duct cell line. Our results revealed major differences in the N-glycosylation of the different PDAC cells as well as compared to the control cell line, suggesting changes of the N-glycosylation in PDAC. The N-glycan profiles of the PDAC cells, however, differed vastly as well and demonstrate the diversity of PDAC model systems, which ultimately affects the interpretation of functional studies. The results from this study form the basis for further biological evaluation of the role of protein glycosylation in PDAC and highlight that conclusions from one cell line cannot be generalised, but should be regarded in the context of the corresponding phenotype.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA: a cancer journal for clinicians. 2016;66:7–30. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

