Nitrogen use efficiency is regulated by interacting proteins relevant to development in wheat
- PMID: 29193541
- PMCID: PMC5978868
- DOI: 10.1111/pbi.12864
Nitrogen use efficiency is regulated by interacting proteins relevant to development in wheat
Abstract
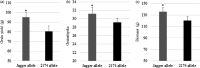
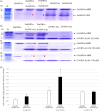
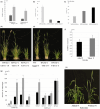
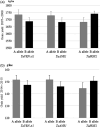
Wheat (Triticum aestivum) has low nitrogen use efficiency (NUE). The genetic mechanisms controlling NUE are unknown. Positional cloning of a major quantitative trait locus for N-related agronomic traits showed that the vernalization gene TaVRN-A1 was tightly linked with TaNUE1, the gene shown to influence NUE in wheat. Because of an Ala180 /Val180 substitution, TaVRN-A1a and TaVRN-A1b proteins interact differentially with TaANR1, a protein encoded by a wheat orthologue of Arabidopsis nitrate regulated 1 (ANR1). The transcripts of both TaVRN-A1 and TaANR1 were down-regulated by nitrogen. TaANR1 was functionally characterized in TaANR1::RNAi transgenic wheat, and in a natural mutant with a 23-bp deletion including 10-bp at the 5' end of intron 5 and 13-bp of exon 6 in gDNA sequence in its gDNA sequence, which produced transcript that lacked the full 84-bp exon 6. Both TaANR1 and TaHOX1 bound to the Ala180 /Val180 position of TaVRN-A1. Genetically incorporating favourable alleles from TaVRN-A1, TaANR1 and TaHOX1 increased grain yield from 9.84% to 11.58% in the field. Molecular markers for allelic variation of the genes that regulate nitrogen can be used in breeding programmes aimed at improving NUE and yield in novel wheat cultivars.
Keywords: TaANR1; TaHOX1; TaVRN-A1; flowering time; nitrogen use efficiency (NUE); wheat.
© 2017 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- An, D. , Su, J. , Liu, Q. , Zhu, Y. , Tong, Y. , Li, J. , Li, J. et al. (2006) Mapping QTLs for nitrogen uptake in relation to the early growth of wheat (Triticum aestivum L.). Plant Soil, 284, 73–84.
-
- Chen, Y. , Carver, B.F. , Wang, S. , Zhang, F. and Yan, L. (2009) Genetic loci associated with stem elongation and winter dormancy release in wheat. Theor. Appl. Genet. 118, 881–889. - PubMed
-
- Cormier, F. , Faure, S. , Dubreuil, P. , Heumez, E. , Beauchêne, K. , Lafarge, S. , Praud, S. et al. (2013) A multi‐environmental study of recent breeding progress on nitrogen use efficiency in wheat (Triticum aestivum L.). Theor. Appl. Genet. 126, 3035–3048. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

