Genetic Fine-Mapping and Identification of Candidate Genes and Variants for Adiposity Traits in Outbred Rats
- PMID: 29193816
- PMCID: PMC5740008
- DOI: 10.1002/oby.22075
Genetic Fine-Mapping and Identification of Candidate Genes and Variants for Adiposity Traits in Outbred Rats
Abstract
Objective: Obesity is a major risk factor for multiple diseases and is in part heritable, yet the majority of causative genetic variants that drive excessive adiposity remain unknown. Here, outbred heterogeneous stock (HS) rats were used in controlled environmental conditions to fine-map novel genetic modifiers of adiposity.
Methods: Body weight and visceral fat pad weights were measured in male HS rats that were also genotyped genome-wide. Quantitative trait loci (QTL) were identified by genome-wide association of imputed single-nucleotide polymorphism (SNP) genotypes using a linear mixed effect model that accounts for unequal relatedness between the HS rats. Candidate genes were assessed by protein modeling and mediation analysis of expression for coding and noncoding variants, respectively.
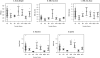
Results: HS rats exhibited large variation in adiposity traits, which were highly heritable and correlated with metabolic health. Fine-mapping of fat pad weight and body weight revealed three QTL and prioritized five candidate genes. Fat pad weight was associated with missense SNPs in Adcy3 and Prlhr and altered expression of Krtcap3 and Slc30a3, whereas Grid2 was identified as a candidate within the body weight locus.
Conclusions: These data demonstrate the power of HS rats for identification of known and novel heritable mediators of obesity traits.
© 2017 The Obesity Society.
Conflict of interest statement
Figures






References
-
- Wang YC, et al. Health and economic burden of the projected obesity trends in the USA and the UK. Lancet. 2011;378(9793):815–25. - PubMed
-
- Stunkard AJ, Foch TT, Hrubec Z. A twin study of human obesity. Jama. 1986;256(1):51–4. - PubMed
-
- Katzmarzyk PT, et al. Familial resemblance in fatness and fat distribution. Am J Hum Biol. 2000;12(3):395–404. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

