Somatic mosaicism of an intragenic FANCB duplication in both fibroblast and peripheral blood cells observed in a Fanconi anemia patient leads to milder phenotype
- PMID: 29193904
- PMCID: PMC5823675
- DOI: 10.1002/mgg3.350
Somatic mosaicism of an intragenic FANCB duplication in both fibroblast and peripheral blood cells observed in a Fanconi anemia patient leads to milder phenotype
Abstract
Background: Fanconi anemia (FA) is a rare disorder characterized by congenital malformations, progressive bone marrow failure, and predisposition to cancer. Patients harboring X-linked FANCB pathogenic variants usually present with severe congenital malformations resembling VACTERL syndrome with hydrocephalus.
Methods: We employed the diepoxybutane (DEB) test for FA diagnosis, arrayCGH for detection of duplication, targeted capture and next-gen sequencing for defining the duplication breakpoint, PacBio sequencing of full-length FANCB aberrant transcript, FANCD2 ubiquitination and foci formation assays for the evaluation of FANCB protein function by viral transduction of FANCB-null cells with lentiviral FANCB WT and mutant expression constructs, and droplet digital PCR for quantitation of the duplication in the genomic DNA and cDNA.
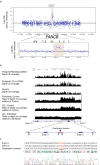
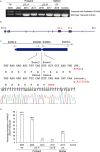
Results: We describe here an FA-B patient with a mild phenotype. The DEB diagnostic test for FA revealed somatic mosaicism. We identified a 9154 bp intragenic duplication in FANCB, covering the first coding exon 3 and the flanking regions. A four bp homology (GTAG) present at both ends of the breakpoint is consistent with microhomology-mediated duplication mechanism. The duplicated allele gives rise to an aberrant transcript containing exon 3 duplication, predicted to introduce a stop codon in FANCB protein (p.A319*). Duplication levels in the peripheral blood DNA declined from 93% to 7.9% in the span of eleven years. Moreover, the patient fibroblasts have shown 8% of wild-type (WT) allele and his carrier mother showed higher than expected levels of WT allele (79% vs. 50%) in peripheral blood, suggesting that the duplication was highly unstable.
Conclusion: Unlike sequence point variants, intragenic duplications are difficult to precisely define, accurately quantify, and may be very unstable, challenging the proper diagnosis. The reversion of genomic duplication to the WT allele results in somatic mosaicism and may explain the relatively milder phenotype displayed by the FA-B patient described here.
Keywords: FANCB; droplet digital PCR; intragenic duplication; milder phenotype; revertant mosaicism.
© 2017 The Authors. Molecular Genetics & Genomic Medicine published by Wiley Periodicals, Inc.
Figures




References
-
- Auerbach, A. D. (2009). Fanconi anemia and its diagnosis. Mutation Research, 668(1–2), 4–10. https://doi.org/10.1016/j.mrfmmm.2009.01.013 - DOI - PMC - PubMed
-
- Biesecker, L. G. , & Spinner, N. B. (2013). A genomic view of mosaicism and human disease. Nature Reviews Genetics, 14(5), 307–320. https://doi.org/10.1038/nrg3424 - DOI - PubMed
-
- Chandrasekharappa, S. C. , Lach, F. P. , Kimble, D. C. , Kamat, A. , Teer, J. K. , Donovan, F. X. , … Smogorzewska, A . (2013). Massively parallel sequencing, aCGH, and RNA‐Seq technologies provide a comprehensive molecular diagnosis of Fanconi anemia. Blood, 121(22), e138–e148. https://www.ncbi.nlm.nih.gov/books/NBK1401/. https://doi.org/10.1182/blood-2012-12-474585 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

