Genome Context Viewer: visual exploration of multiple annotated genomes using microsynteny
- PMID: 29194466
- PMCID: PMC5925771
- DOI: 10.1093/bioinformatics/btx757
Genome Context Viewer: visual exploration of multiple annotated genomes using microsynteny
Abstract
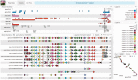
Summary: The Genome Context Viewer is a visual data-mining tool that allows users to search across multiple providers of genome data for regions with similarly annotated content that may be aligned and visualized at the level of their shared functional elements. By handling ordered sequences of gene family memberships as a unit of search and comparison, the user interface enables quick and intuitive assessment of the degree of gene content divergence and the presence of various types of structural events within syntenic contexts. Insights into functionally significant differences seen at this level of abstraction can then serve to direct the user to more detailed explorations of the underlying data in other interconnected, provider-specific tools.
Availability and implementation: GCV is provided under the GNU General Public License version 3 (GPL-3.0). Source code is available at https://github.com/legumeinfo/lis_context_viewer.
Contact: adf@ncgr.org.
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures
References
-
- Dash S. et al. (2016) Legume information system (legumeinfo.org): a key component of a set of federated data resources for the legume family. Nucleic Acids Research, 44, D1181–D1188. - PMC - PubMed
-
- Durbin R. et al. (1998) Biological Sequence Analysis: probabilistic Models of Proteins and Nucleic Acids. Cambridge University Press, Cambridge.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

