bioBakery: a meta'omic analysis environment
- PMID: 29194469
- PMCID: PMC6030947
- DOI: 10.1093/bioinformatics/btx754
bioBakery: a meta'omic analysis environment
Abstract
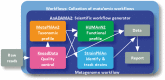
Summary: bioBakery is a meta'omic analysis environment and collection of individual software tools with the capacity to process raw shotgun sequencing data into actionable microbial community feature profiles, summary reports, and publication-ready figures. It includes a collection of pre-configured analysis modules also joined into workflows for reproducibility.
Availability and implementation: bioBakery (http://huttenhower.sph.harvard.edu/biobakery) is publicly available for local installation as individual modules and as a virtual machine image. Each individual module has been developed to perform a particular task (e.g. quantitative taxonomic profiling or statistical analysis), and they are provided with source code, tutorials, demonstration data, and validation results; the bioBakery virtual image includes the entire suite of modules and their dependencies pre-installed. Images are available for both Amazon EC2 and Google Compute Engine. All software is open source under the MIT license. bioBakery is actively maintained with a support group at biobakery-users@googlegroups.com and new tools being added upon their release.
Contact: chuttenh@hsph.harvard.edu.
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources