Shotgun Metagenomics and Volatilome Profile of the Microbiota of Fermented Sausages
- PMID: 29196291
- PMCID: PMC5772244
- DOI: 10.1128/AEM.02120-17
Shotgun Metagenomics and Volatilome Profile of the Microbiota of Fermented Sausages
Abstract
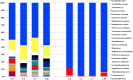
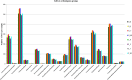
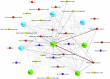
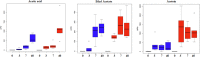
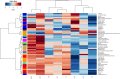
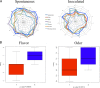
Changes in the microbial gene content and abundance can be analyzed to detect shifts in the microbiota composition due to the use of a starter culture in the food fermentation process, with the consequent shift of key metabolic pathways directly connected with product acceptance. Meat fermentation is a complex process involving microbes that metabolize the main components in meat. The breakdown of carbohydrates, proteins, and lipids can lead to the formation of volatile organic compounds (VOCs) that can drastically affect the organoleptic characteristics of the final products. The present meta-analysis, performed with the shotgun DNA metagenomic approach, focuses on studying the microbiota and its gene content in an Italian fermented sausage produced by using a commercial starter culture (a mix of Lactobacillus sakei and Staphylococcus xylosus), with the aim to discover the connections between the microbiota, microbiome, and the release of volatile metabolites during ripening. The inoculated fermentation with the starter culture limited the development of Enterobacteriaceae and reduced the microbial diversity compared to that from spontaneous fermentation. KEGG database genes associated with the reduction of acetaldehyde to ethanol (EC 1.1.1.1), acetyl phosphate to acetate (EC 2.7.2.1), and 2,3-butanediol to acetoin (EC 1.1.1.4) were most abundant in inoculated samples (I) compared to those in spontaneous fermentation samples (S). The volatilome profiles were highly consistent with the abundance of the genes; elevated acetic acid (1,173.85 μg/kg), ethyl acetate (251.58 μg/kg), and acetoin (1,100.19 μg/kg) were observed in the presence of the starters at the end of fermentation. Significant differences were found in the liking of samples based on flavor and odor, suggesting a higher preference by consumers for the spontaneous fermentation samples. Inoculated samples exhibited the lowest scores for the liking data, which were clearly associated with the highest concentration of acetic acid.IMPORTANCE We present an advance in the understanding of meat fermentation by coupling DNA sequencing metagenomics and metabolomics approaches to describe the microbial function during this process. Very few studies using this global approach have been dedicated to food, and none have examined sausage fermentation, underlying the originality of the study. The starter culture drastically affected the organoleptic properties of the products. This finding underlines the importance of starter culture selection that takes into consideration the functional characteristics of the microorganism to optimize production efficiency and product quality.
Keywords: fermented sausages; metabolic pathways; shotgun metagenomics; volatile organic compounds.
Copyright © 2018 American Society for Microbiology.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

