Quasimetagenomics-Based and Real-Time-Sequencing-Aided Detection and Subtyping of Salmonella enterica from Food Samples
- PMID: 29196295
- PMCID: PMC5795075
- DOI: 10.1128/AEM.02340-17
Quasimetagenomics-Based and Real-Time-Sequencing-Aided Detection and Subtyping of Salmonella enterica from Food Samples
Abstract
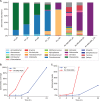
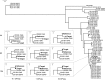
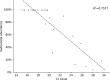
Metagenomics analysis of food samples promises isolation-independent detection and subtyping of foodborne bacterial pathogens in a single workflow. The selective concentration of Salmonella genomic DNA by immunomagnetic separation (IMS) and multiple displacement amplification (MDA) shortened the time for culture enrichment of Salmonella-spiked raw chicken breast samples by over 12 h while permitting serotyping and high-fidelity single nucleotide polymorphism (SNP) typing of the pathogen using short shotgun sequencing reads. The herein-termed quasimetagenomics approach was evaluated on Salmonella-spiked lettuce and black peppercorn samples as well as retail chicken parts naturally contaminated with different serotypes of Salmonella Culture enrichment of between 8 and 24 h was required for detecting and subtyping naturally occurring Salmonella from unspiked chicken parts compared with 4- to 12-h culture enrichment when Salmonella-spiked food samples were analyzed, indicating the likely need for longer culture enrichment to revive low levels of stressed or injured Salmonella cells in food. A further acceleration of the workflow was achieved by real-time nanopore sequencing. After 1.5 h of analysis on a potable sequencer, sufficient data were generated from sequencing the IMS-MDA products of a cultured-enriched lettuce sample to enable serotyping and robust phylogenetic placement of the inoculated isolate.IMPORTANCE Both culture enrichment and next-generation sequencing remain time-consuming processes for food testing, whereas rapid methods for pathogen detection are widely available. Our study demonstrated a substantial acceleration of these processes by the use of immunomagnetic separation (IMS) with multiple displacement amplification (MDA) and real-time nanopore sequencing. In one example, the combined use of the two methods delivered a less than 24-h turnaround time from the collection of a Salmonella-contaminated lettuce sample to the phylogenetic identification of the pathogen. An improved efficiency such as this is important for further expanding the use of whole-genome and metagenomics sequencing in the microbial analysis of food. Our results suggest the potential of the quasimetagenomics approach in areas where rapid detection and subtyping of foodborne pathogens are important, such as for foodborne outbreak response and the precision tracking and monitoring of foodborne pathogens in production environments and supply chains.
Keywords: MinION; Salmonella; detection; metagenomics; subtyping.
Copyright © 2018 American Society for Microbiology.
Figures
References
-
- Deng X, Shariat N, Driebe EM, Roe CC, Tolar B, Trees E, Keim P, Zhang W, Dudley EG, Fields PI, Engelthaler DM. 2015. Comparative analysis of subtyping methods against a whole-genome-sequencing standard for Salmonella enterica serotype Enteritidis. J Clin Microbiol 53:212–218. doi: 10.1128/JCM.02332-14. - DOI - PMC - PubMed
-
- Moura A, Criscuolo A, Pouseele H, Maury MM, Leclercq A, Tarr C, Bjorkman JT, Dallman T, Reimer A, Enouf V, Larsonneur E, Carleton H, Bracq-Dieye H, Katz LS, Jones L, Touchon M, Tourdjman M, Walker M, Stroika S, Cantinelli T, Chenal-Francisque V, Kucerova Z, Rocha EP, Nadon C, Grant K, Nielsen EM, Pot B, Gerner-Smidt P, Lecuit M, Brisse S. 2016. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat Microbiol 2:16185. doi: 10.1038/nmicrobiol.2016.185. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

