A small cassette enables conditional gene inactivation by CRISPR/Cas9
- PMID: 29196747
- PMCID: PMC5711947
- DOI: 10.1038/s41598-017-16931-z
A small cassette enables conditional gene inactivation by CRISPR/Cas9
Abstract
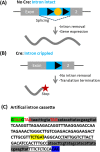
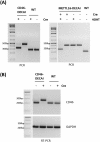
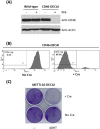
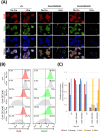
The availability of CRISPR/Cas9 technology has enabled the rapid establishment of gene knockouts in many cell types and even whole organisms. However, conditional inactivation of essential genes remains a challenge. We devised an approach named DECAI (DEgradation based on Cre-regulated- Artificial Intron). It utilizes a small cassette of just 201 nucleotides that is inserted into the coding exon of a target gene using CRISPR/Cas9 technology and homology-directed repair. As its sequence is derived from an artificial intron, the cassette is removed by the splicing machinery and thus leaves no trace in the "off-state". Upon activation with Cre recombinase ("on-state"), the intron is crippled and the target gene is disrupted by a series of stop codons. We exemplify the utility of this approach on several non-essential and essential human genes. Clones bearing the conditional knockout cassette are recovered at frequencies above 5% and cassette function can be traced at the genomic DNA and the mRNA level. Importantly, cassette activation leads to loss of gene expression as judged by flow cytometry, Western blot or immunofluorescence. Altogether, this highlights the broad utility of the approach for conditional gene inactivation and suggests that this tool could be used to study the loss-of-function phenotypes of essential genes.
Conflict of interest statement
Paloma Guzzardo, Christina Rashkova and Tilmann Buerckstuemmer are employees of Horizon Genomics GmbH. Rodrigo L. dos Santos and Philippe Collin are employees of Horizon Discover Ltd. Horizon has filed a UK provisional patent covering this approach.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

