Whole exome sequencing identifies TRIOBP pathogenic variants as a cause of post-lingual bilateral moderate-to-severe sensorineural hearing loss
- PMID: 29197352
- PMCID: PMC5712175
- DOI: 10.1186/s12881-017-0499-z
Whole exome sequencing identifies TRIOBP pathogenic variants as a cause of post-lingual bilateral moderate-to-severe sensorineural hearing loss
Abstract
Background: Implementation of whole exome sequencing has provided unique opportunity for a wide screening of causative variants in genetically heterogeneous diseases, including nonsyndromic hearing impairment. TRIOBP in the inner ear is responsible for proper structure and function of stereocilia and is necessary for sound transduction.
Methods: Whole exome sequencing followed by Sanger sequencing was conducted on patients derived from Polish hearing loss family.
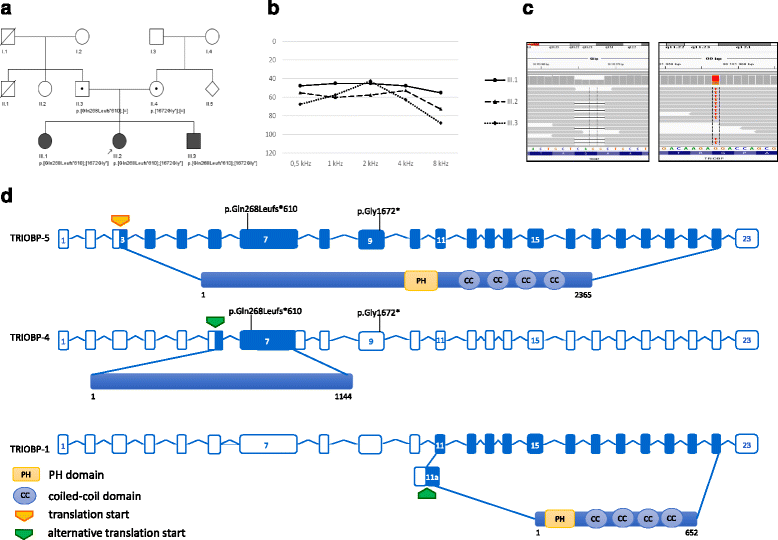
Results: Based on whole exome analysis, we identified two TRIOBP pathogenic variants (c.802_805delCAGG, p.Gln268Leufs*610 and c.5014G>T, p.Gly1672*, the first of which was novel) causative of nonsyndromic, peri- to postlingual, moderate-to-severe hearing loss in three siblings from a Polish family. Typically, TRIOBP pathogenic variants lead to prelingual, severe-to-profound hearing loss, thus the onset and degree of hearing impairment in our patients represent a distinct phenotypic manifestation caused by TRIOBP variants. The pathogenic variant p.Gln268Leufs*610 disrupts the TRIOBP-4 and TRIOBP-5 isoforms (both expressed exclusively in the inner ear and retina) whereas the second pathogenic variant c.514G>T, p.Gly1672* affects only TRIOBP-5.
Conclusions: The onset and degree of hearing impairment, characteristic for our patients, represent a unique phenotypic manifestation caused by TRIOBP pathogenic variants. Although TRIOBP alterations are not a frequent cause of hearing impairment, this gene should be thoroughly analyzed especially in patients with a postlingual hearing loss. A delayed onset of hearing impairment due to TRIOBP pathogenic variants creates a potential therapeutic window for future targeted therapies.
Keywords: Hearing impairment; TRIOBP; Whole exome sequencing.
Conflict of interest statement
Ethics approval and consent to participate
The study protocol was approved by Ethical Committee of the Institute of Physiology and Pathology of Hearing (reference number IFPS:/KB/03/2012). Patients or guardians provided informed written consent prior to participation.
Consent for publication
The patients or their guardians gave written consent for the publication of their medical information.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- Diaz-Horta O, Duman D, Foster J, 2nd, Sirmaci A, Gonzalez M, Mahdieh N, Fotouhi N, Bonyadi M, Cengiz FB, Menendez I, et al. Whole-exome sequencing efficiently detects rare mutations in autosomal recessive nonsyndromic hearing loss. PLoS One. 2012;7(11):e50628. doi: 10.1371/journal.pone.0050628. - DOI - PMC - PubMed
-
- Seipel K, O'Brien SP, Iannotti E, Medley QG, Streuli M. Tara, a novel F-actin binding protein, associates with the trio guanine nucleotide exchange factor and regulates actin cytoskeletal organization. J Cell Sci. 2001;114(Pt 2):389–399. - PubMed
-
- Hirosawa M, Nagase T, Murahashi Y, Kikuno R, Ohara O. Identification of novel transcribed sequences on human chromosome 22 by expressed sequence tag mapping. DNA research: an international journal for rapid publication of reports on genes and genomes. 2001;8(1):1–9. doi: 10.1093/dnares/8.1.1. - DOI - PubMed
-
- Riazuddin S, Khan SN, Ahmed ZM, Ghosh M, Caution K, Nazli S, Kabra M, Zafar AU, Chen K, Naz S, et al. Mutations in TRIOBP, which encodes a putative cytoskeletal-organizing protein, are associated with nonsyndromic recessive deafness. Am J Hum Genet. 2006;78(1):137–143. doi: 10.1086/499164. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

