Expansion of the Genetic Alphabet: A Chemist's Approach to Synthetic Biology
- PMID: 29198111
- PMCID: PMC5820176
- DOI: 10.1021/acs.accounts.7b00403
Expansion of the Genetic Alphabet: A Chemist's Approach to Synthetic Biology
Abstract
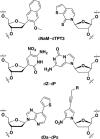
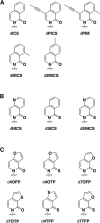
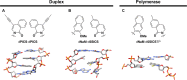
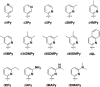
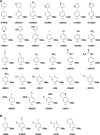
The information available to any organism is encoded in a four nucleotide, two base pair genetic code. Since its earliest days, the field of synthetic biology has endeavored to impart organisms with novel attributes and functions, and perhaps the most fundamental approach to this goal is the creation of a fifth and sixth nucleotide that pair to form a third, unnatural base pair (UBP) and thus allow for the storage and retrieval of increased information. Achieving this goal, by definition, requires synthetic chemistry to create unnatural nucleotides and a medicinal chemistry-like approach to guide their optimization. With this perspective, almost 20 years ago we began designing unnatural nucleotides with the ultimate goal of developing UBPs that function in vivo, and thus serve as the foundation of semi-synthetic organisms (SSOs) capable of storing and retrieving increased information. From the beginning, our efforts focused on the development of nucleotides that bear predominantly hydrophobic nucleobases and thus that pair not based on the complementary hydrogen bonds that are so prominent among the natural base pairs but rather via hydrophobic and packing interactions. It was envisioned that such a pairing mechanism would provide a basal level of selectivity against pairing with natural nucleotides, which we expected would be the greatest challenge; however, this choice mandated starting with analogs that have little or no homology to their natural counterparts and that, perhaps not surprisingly, performed poorly. Progress toward their optimization was driven by the construction of structure-activity relationships, initially from in vitro steady-state kinetic analysis, then later from pre-steady-state and PCR-based assays, and ultimately from performance in vivo, with the results augmented three times with screens that explored combinations of the unnatural nucleotides that were too numerous to fully characterize individually. The structure-activity relationship data identified multiple features required by the UBP, and perhaps most prominent among them was a substituent ortho to the glycosidic linkage that is capable of both hydrophobic packing and hydrogen bonding, and nucleobases that stably stack with flanking natural nucleobases in lieu of the potentially more stabilizing stacking interactions afforded by cross strand intercalation. Most importantly, after the examination of hundreds of unnatural nucleotides and thousands of candidate UBPs, the efforts ultimately resulted in the identification of a family of UBPs that are well recognized by DNA polymerases when incorporated into DNA and that have been used to create SSOs that store and retrieve increased information. In addition to achieving a longstanding goal of synthetic biology, the results have important implications for our understanding of both the molecules and forces that can underlie biological processes, so long considered the purview of molecules benefiting from eons of evolution, and highlight the promise of applying the approaches and methodologies of synthetic and medical chemistry in the pursuit of synthetic biology.
Conflict of interest statement
The authors declare the following competing financial interest(s): F.E.R. has a financial interest (shares) in Synthorx Inc., a company that has commercial interests in the UBP. The other author declares no other competing financial interests.
Figures








References
-
- Leduc S.The Mechanisms of Life; Rebman Company: New York, 1911.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

