Stress-response transcription factors Msn2 and Msn4 couple TORC2-Ypk1 signaling and mitochondrial respiration to ATG8 gene expression and autophagy
- PMID: 29198169
- PMCID: PMC5788474
- DOI: 10.1080/15548627.2017.1356949
Stress-response transcription factors Msn2 and Msn4 couple TORC2-Ypk1 signaling and mitochondrial respiration to ATG8 gene expression and autophagy
Abstract
Macroautophagy/autophagy is a starvation and stress-induced catabolic process critical for cellular homeostasis and adaptation. Several Atg proteins are involved in the formation of the autophagosome and subsequent degradation of cytoplasmic components, a process termed autophagy flux. Additionally, the expression of several Atg proteins, in particular Atg8, is modulated transcriptionally, yet the regulatory mechanisms involved remain poorly understood. Here we demonstrate that the AGC kinase Ypk1, target of the rapamycin-insensitive TORC2 signaling pathway, controls ATG8 expression by repressing the heterodimeric Zinc-finger transcription factors Msn2 and Msn4. We find that Msn2 and Msn4 promote ATG8 expression downstream of the histone deacetylase complex (HDAC) subunit Ume6, a previously identified negative regulator of ATG8 expression. Moreover, we demonstrate that TORC2-Ypk1 signaling is functionally linked to distinct mitochondrial respiratory complexes. Surprisingly, we find that autophagy flux during amino acid starvation is also dependent upon Msn2-Msn4 activity, revealing a broad role for these transcription factors in the autophagy response.
Keywords: TOR signaling; Ypk1; autophagy; electron transport chain (ETC) complexes; gene expression; mitochondrial respiration; signal transduction.
Figures
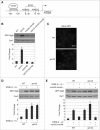
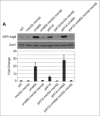
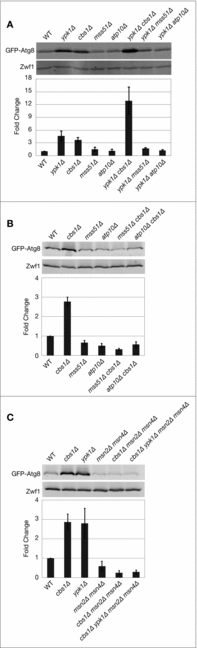


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
