Bone Marrow Myeloid Cells Regulate Myeloid-Biased Hematopoietic Stem Cells via a Histamine-Dependent Feedback Loop
- PMID: 29198940
- PMCID: PMC5975960
- DOI: 10.1016/j.stem.2017.11.003
Bone Marrow Myeloid Cells Regulate Myeloid-Biased Hematopoietic Stem Cells via a Histamine-Dependent Feedback Loop
Abstract
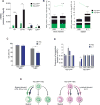
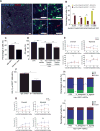
Myeloid-biased hematopoietic stem cells (MB-HSCs) play critical roles in recovery from injury, but little is known about how they are regulated within the bone marrow niche. Here we describe an auto-/paracrine physiologic circuit that controls quiescence of MB-HSCs and hematopoietic progenitors marked by histidine decarboxylase (Hdc). Committed Hdc+ myeloid cells lie in close anatomical proximity to MB-HSCs and produce histamine, which activates the H2 receptor on MB-HSCs to promote their quiescence and self-renewal. Depleting histamine-producing cells enforces cell cycle entry, induces loss of serial transplant capacity, and sensitizes animals to chemotherapeutic injury. Increasing demand for myeloid cells via lipopolysaccharide (LPS) treatment specifically recruits MB-HSCs and progenitors into the cell cycle; cycling MB-HSCs fail to revert into quiescence in the absence of histamine feedback, leading to their depletion, while an H2 agonist protects MB-HSCs from depletion after sepsis. Thus, histamine couples lineage-specific physiological demands to intrinsically primed MB-HSCs to enforce homeostasis.
Keywords: H2 receptor; bone marrow niche; hematopoietic stem cells; histamine; histidine decarboxylase; myeloid biased; quiescence; self-renewal.
Published by Elsevier Inc.
Figures




Comment in
-
Saving Stem Cells after Stress.Cell Stem Cell. 2017 Dec 7;21(6):709-710. doi: 10.1016/j.stem.2017.11.018. Cell Stem Cell. 2017. PMID: 29220661
References
-
- Alcaniz L, Vega A, Chacon P, El Bekay R, Ventura I, Aroca R, Blanca M, Bergstralh DT, Monteseirin J. Histamine production by human neutrophils. FASEB J. 2013;27:2902–2910. - PubMed
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate; A practical and powerful approach to multiple testing. J Roy Stat Soc Ser B. 1995;57:289–300.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

