Reverse engineering the cancer metabolic network using flux analysis to understand drivers of human disease
- PMID: 29199104
- PMCID: PMC5927620
- DOI: 10.1016/j.ymben.2017.11.013
Reverse engineering the cancer metabolic network using flux analysis to understand drivers of human disease
Abstract
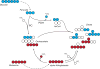
Metabolic dysfunction has reemerged as an essential hallmark of tumorigenesis, and metabolic phenotypes are increasingly being integrated into pre-clinical models of disease. The complexity of these metabolic networks requires systems-level interrogation, and metabolic flux analysis (MFA) with stable isotope tracing present a suitable conceptual framework for such systems. Here we review efforts to elucidate mechanisms through which metabolism influences tumor growth and survival, with an emphasis on applications using stable isotope tracing and MFA. Through these approaches researchers can now quantify pathway fluxes in various in vitro and in vivo contexts to provide mechanistic insights at molecular and physiological scales respectively. Knowledge and discoveries in cancer models are paving the way toward applications in other biological contexts and disease models. In turn, MFA approaches will increasingly help to uncover new therapeutic opportunities that enhance human health.
Keywords: Cancer; Metabolic flux analysis; Metabolism; Metabolomics; Mitochondria; Stable isotope tracing.
Copyright © 2017 International Metabolic Engineering Society. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Antoniewicz MR, Kelleher JK, Stephanopoulos G. Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metab Eng. 2006;8:324–37. - PubMed
-
- Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116:597–602. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

