STAT5BN642H is a driver mutation for T cell neoplasia
- PMID: 29200404
- PMCID: PMC5749501
- DOI: 10.1172/JCI94509
STAT5BN642H is a driver mutation for T cell neoplasia
Abstract
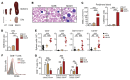
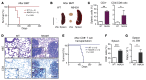
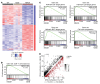
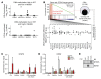
STAT5B is often mutated in hematopoietic malignancies. The most frequent STAT5B mutation, Asp642His (N642H), has been found in over 90 leukemia and lymphoma patients. Here, we used the Vav1 promoter to generate transgenic mouse models that expressed either human STAT5B or STAT5BN642H in the hematopoietic compartment. While STAT5B-expressing mice lacked a hematopoietic phenotype, the STAT5BN642H-expressing mice rapidly developed T cell neoplasms. Neoplasia manifested as transplantable CD8+ lymphoma or leukemia, indicating that the STAT5BN642H mutation drives cancer development. Persistent and enhanced levels of STAT5BN642H tyrosine phosphorylation in transformed CD8+ T cells led to profound changes in gene expression that were accompanied by alterations in DNA methylation at potential histone methyltransferase EZH2-binding sites. Aurora kinase genes were enriched in STAT5BN642H-expressing CD8+ T cells, which were exquisitely sensitive to JAK and Aurora kinase inhibitors. Together, our data suggest that JAK and Aurora kinase inhibitors should be further explored as potential therapeutics for lymphoma and leukemia patients with the STAT5BN642H mutation who respond poorly to conventional chemotherapy.
Keywords: Hematology; Leukemias; Lymphomas; Oncology; T cells.
Conflict of interest statement
Figures




Comment in
-
Rare mutations provide unique insight into oncogenic potential of STAT transcription factors.J Clin Invest. 2018 Jan 2;128(1):113-115. doi: 10.1172/JCI98619. Epub 2017 Dec 4. J Clin Invest. 2018. PMID: 29199995 Free PMC article.
References
-
- Nivarthi H, Friedbichler K, Moriggl R. Stat5 as a Hematopoietic Master Regulator for Differentiation and Neoplasia Development. In: Decker T, Müller M, eds. Jak-Stat Signaling: From Basics to Disease. Vienna: Springer;2012:153–167.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

