Transcriptomic Response of Resistant (PI613981- Malus sieversii) and Susceptible ("Royal Gala") Genotypes of Apple to Blue Mold (Penicillium expansum) Infection
- PMID: 29201037
- PMCID: PMC5696741
- DOI: 10.3389/fpls.2017.01981
Transcriptomic Response of Resistant (PI613981- Malus sieversii) and Susceptible ("Royal Gala") Genotypes of Apple to Blue Mold (Penicillium expansum) Infection
Abstract
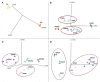
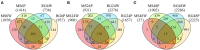
Malus sieversii from Central Asia is a progenitor of the modern domesticated apple (Malus × domestica). Several accessions of M. sieversii are highly resistant to the postharvest pathogen Penicillium expansum. A previous study identified the qM-Pe3.1 QTL on LG3 for resistance to P. expansum in the mapping population GMAL4593, developed using the resistant accession, M. sieversii -PI613981, and the susceptible cultivar "Royal Gala" (RG) (M. domestica), as parents. The goal of the present study was to characterize the transcriptomic response of susceptible RG and resistant PI613981 apple fruit to wounding and inoculation with P. expansum using RNA-Seq. Transcriptomic analyses 0-48 h post inoculation suggest a higher basal level of resistance and a more rapid and intense defense response to wounding and wounding plus inoculation with P. expansum in M. sieversii -PI613981 than in RG. Functional analysis showed that ethylene-related genes and genes involved in "jasmonate" and "MYB-domain transcription factor family" were over-represented in the resistant genotype. It is suggested that the more rapid response in the resistant genotype (Malus sieversii-PI613981) plays a major role in the resistance response. At least twenty DEGs were mapped to the qM-Pe3.1 QTL (M × d v.1: 26,848,396-28,424,055) on LG3, and represent potential candidate genes responsible for the observed resistance QTL in M. sieversii-PI613981. RT-qPCR of several of these genes was used to validate the RNA-Seq data and to confirm their higher expression in MS0.
Keywords: Penicillium expansum; malus domestica; necrotrophic pathogens; postharvest pathogens; qualitative trait loci; wound healing.
Figures




References
-
- Ahmadi-Afzadi M., Nybom H., Ekholm A., Tahir I., Rumpunen K. (2015). Biochemical contents of apple peel and flesh affect level of partial resistance to blue mold. Postharvest Biol. Technol. 110, 173–182. 10.1016/j.postharvbio.2015.08.008 - DOI
-
- Ahmadi-Afzadi M., Tahir I., Nybom H. (2013). Impact of harvesting time and fruit firmness on the tolerance to fungal storage diseases in an apple germplasm collection. Postharvest Biol. Technol. 82, 51–58. 10.1016/j.postharvbio.2013.03.001 - DOI
-
- Andersen C. L., Jensen J. L., Ørntoft T. F. (2004). Normalization of real-ime quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. 10.1158/0008-5472.CAN-04-0496 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

