tRNAs as primers and inhibitors of retrotransposons
- PMID: 29201533
- PMCID: PMC5703155
- DOI: 10.1080/2159256X.2017.1393490
tRNAs as primers and inhibitors of retrotransposons
Abstract
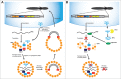
The functional relationship between tRNAs and retrotransposons have been known for more than 35 years. tRNAs are used as primer molecules to guide the reverse transcription of retrotransposons. Recently, tRNAs have also emerge as important players in the postranscriptional regulation of retrotransposons by means of tRNA-derived small RNAs. This surprisingly new layer of regulation indicates that tRNAs are used both in the promotion and the suppression of the reverse transcription of retrotransposons indicating their primary role in the life cycle of LTR retrotransposons. This adds another level of translational control to tRNAs. Here we review the different known levels of interactions of tRNAs and retrotransposons and highlight the unknown parts of this interaction.
Keywords: RNAi; retrotransposons; small RNAs; tRNA-derived fragments; tRNAs.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
