Predictable, Tunable Protein Production in Salmonella for Studying Host-Pathogen Interactions
- PMID: 29201859
- PMCID: PMC5696353
- DOI: 10.3389/fcimb.2017.00475
Predictable, Tunable Protein Production in Salmonella for Studying Host-Pathogen Interactions
Abstract
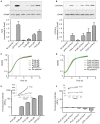
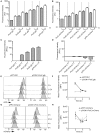
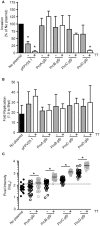
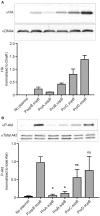
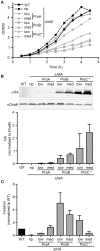
Here we describe the use of synthetic genetic elements to improve the predictability and tunability of episomal protein production in Salmonella. We used a multi-pronged approach, in which a series of variable-strength synthetic promoters were combined with a synthetic transcriptional terminator, and plasmid copy number variation. This yielded a series of plasmids that drive uniform production of fluorescent and endogenous proteins, over a wide dynamic range. We describe several examples where this system is used to fine-tune constitutive expression in Salmonella, providing an efficient means to titrate out toxic effects of protein production.
Keywords: Salmonella; fluorescent protein; intracellular; plasmid; promoter; synthetic biology.
Figures



Similar articles
-
The flagellar regulator fliT represses Salmonella pathogenicity island 1 through flhDC and fliZ.PLoS One. 2012;7(3):e34220. doi: 10.1371/journal.pone.0034220. Epub 2012 Mar 27. PLoS One. 2012. PMID: 22479568 Free PMC article.
-
Controlled Activity of the Salmonella Invasion-Associated Injectisome Reveals Its Intracellular Role in the Cytosolic Population.mBio. 2017 Dec 5;8(6):e01931-17. doi: 10.1128/mBio.01931-17. mBio. 2017. PMID: 29208746 Free PMC article.
-
The bacterial cytoskeleton modulates motility, type 3 secretion, and colonization in Salmonella.PLoS Pathog. 2012 Jan;8(1):e1002500. doi: 10.1371/journal.ppat.1002500. Epub 2012 Jan 26. PLoS Pathog. 2012. PMID: 22291596 Free PMC article.
-
Regulation of Salmonella-host cell interactions via the ubiquitin system.Int J Med Microbiol. 2018 Jan;308(1):176-184. doi: 10.1016/j.ijmm.2017.11.003. Epub 2017 Nov 6. Int J Med Microbiol. 2018. PMID: 29126744 Review.
-
The virulence plasmids of Salmonella.Int Microbiol. 1999 Sep;2(3):177-84. Int Microbiol. 1999. PMID: 10943411 Review.
Cited by
-
Fluorescent tools for the standardized work in Gram-negative bacteria.J Biol Eng. 2024 Apr 8;18(1):25. doi: 10.1186/s13036-024-00420-9. J Biol Eng. 2024. PMID: 38589953 Free PMC article.
-
Salmonella pathogenicity Island 1 undergoes decay in serovars adapted to swine and poultry.Microbiol Spectr. 2025 Jan 7;13(1):e0264324. doi: 10.1128/spectrum.02643-24. Epub 2024 Dec 11. Microbiol Spectr. 2025. PMID: 39660884 Free PMC article.
-
A role for the Salmonella Type III Secretion System 1 in bacterial adaptation to the cytosol of epithelial cells.Mol Microbiol. 2019 Oct;112(4):1270-1283. doi: 10.1111/mmi.14361. Epub 2019 Aug 18. Mol Microbiol. 2019. PMID: 31370104 Free PMC article.
-
Recent advances in genetic systems in obligate intracellular human-pathogenic bacteria.Front Cell Infect Microbiol. 2023 Jun 19;13:1202245. doi: 10.3389/fcimb.2023.1202245. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37404720 Free PMC article. Review.
-
HilD-regulated chemotaxis proteins contribute to Salmonella Typhimurium colonization in the gut.mBio. 2025 Apr 9;16(4):e0039025. doi: 10.1128/mbio.00390-25. Epub 2025 Feb 25. mBio. 2025. PMID: 39998229 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

