Inhibiting DNA Polymerases as a Therapeutic Intervention against Cancer
- PMID: 29201867
- PMCID: PMC5696574
- DOI: 10.3389/fmolb.2017.00078
Inhibiting DNA Polymerases as a Therapeutic Intervention against Cancer
Abstract
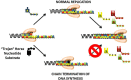
Inhibiting DNA synthesis is an important therapeutic strategy that is widely used to treat a number of hyperproliferative diseases including viral infections, autoimmune disorders, and cancer. This chapter describes two major categories of therapeutic agents used to inhibit DNA synthesis. The first category includes purine and pyrmidine nucleoside analogs that directly inhibit DNA polymerase activity. The second category includes DNA damaging agents including cisplatin and chlorambucil that modify the composition and structure of the nucleic acid substrate to indirectly inhibit DNA synthesis. Special emphasis is placed on describing the molecular mechanisms of these inhibitory effects against chromosomal and mitochondrial DNA polymerases. Discussions are also provided on the mechanisms associated with resistance to these therapeutic agents. A primary focus is toward understanding the roles of specialized DNA polymerases that by-pass DNA lesions produced by DNA damaging agents. Finally, a section is provided that describes emerging areas in developing new therapeutic strategies targeting specialized DNA polymerases.
Keywords: DNA damaging agents; DNA polymerases; cancer; chemotherapy; nucleoside analogs.
Figures






References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

