A Simple and Universal System for Gene Manipulation in Aspergillus fumigatus: In Vitro-Assembled Cas9-Guide RNA Ribonucleoproteins Coupled with Microhomology Repair Templates
- PMID: 29202040
- PMCID: PMC5700375
- DOI: 10.1128/mSphere.00446-17
A Simple and Universal System for Gene Manipulation in Aspergillus fumigatus: In Vitro-Assembled Cas9-Guide RNA Ribonucleoproteins Coupled with Microhomology Repair Templates
Abstract
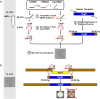
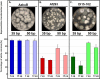
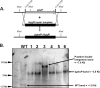
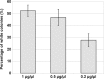
CRISPR (clustered regularly interspaced short palindromic repeat)-Cas9 is a novel genome-editing system that has been successfully established in Aspergillus fumigatus. However, the current state of the technology relies heavily on DNA-based expression cassettes for delivering Cas9 and the guide RNA (gRNA) to the cell. Therefore, the power of the technology is limited to strains that are engineered to express Cas9 and gRNA. To overcome such limitations, we developed a simple and universal CRISPR-Cas9 system for gene deletion that works across different genetic backgrounds of A. fumigatus. The system employs in vitro assembly of dual Cas9 ribonucleoproteins (RNPs) for targeted gene deletion. Additionally, our CRISPR-Cas9 system utilizes 35 to 50 bp of flanking regions for mediating homologous recombination at Cas9 double-strand breaks (DSBs). As a proof of concept, we first tested our system in the ΔakuB (ΔakuBku80 ) laboratory strain and generated high rates (97%) of gene deletion using 2 µg of the repair template flanked by homology regions as short as 35 bp. Next, we inspected the portability of our system across other genetic backgrounds of A. fumigatus, namely, the wild-type strain Af293 and a clinical isolate, A. fumigatus DI15-102. In the Af293 strain, 2 µg of the repair template flanked by 35 and 50 bp of homology resulted in highly efficient gene deletion (46% and 74%, respectively) in comparison to classical gene replacement systems. Similar deletion efficiencies were also obtained in the clinical isolate DI15-102. Taken together, our data show that in vitro-assembled Cas9 RNPs coupled with microhomology repair templates are an efficient and universal system for gene manipulation in A. fumigatus. IMPORTANCE Tackling the multifactorial nature of virulence and antifungal drug resistance in A. fumigatus requires the mechanistic interrogation of a multitude of genes, sometimes across multiple genetic backgrounds. Classical fungal gene replacement systems can be laborious and time-consuming and, in wild-type isolates, are impeded by low rates of homologous recombination. Our simple and universal CRISPR-Cas9 system for gene manipulation generates efficient gene targeting across different genetic backgrounds of A. fumigatus. We anticipate that our system will simplify genome editing in A. fumigatus, allowing for the generation of single- and multigene knockout libraries. In addition, our system will facilitate the delineation of virulence factors and antifungal drug resistance genes in different genetic backgrounds of A. fumigatus.
Keywords: Aspergillus fumigatus; CRISPR-Cas9; gene deletion; genome editing; in vitro assembly; pksP gene.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
