Adaptation in a Fibronectin Binding Autolysin of Staphylococcus saprophyticus
- PMID: 29202045
- PMCID: PMC5705806
- DOI: 10.1128/mSphere.00511-17
Adaptation in a Fibronectin Binding Autolysin of Staphylococcus saprophyticus
Abstract
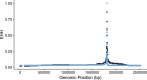
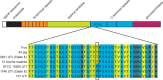
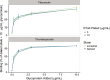
Human-pathogenic bacteria are found in a variety of niches, including free-living, zoonotic, and microbiome environments. Identifying bacterial adaptations that enable invasive disease is an important means of gaining insight into the molecular basis of pathogenesis and understanding pathogen emergence. Staphylococcus saprophyticus, a leading cause of urinary tract infections, can be found in the environment, food, animals, and the human microbiome. We identified a selective sweep in the gene encoding the Aas adhesin, a key virulence factor that binds host fibronectin. We hypothesize that the mutation under selection (aas_2206A>C) facilitates colonization of the urinary tract, an environment where bacteria are subject to strong shearing forces. The mutation appears to have enabled emergence and expansion of a human-pathogenic lineage of S. saprophyticus. These results demonstrate the power of evolutionary genomic approaches in discovering the genetic basis of virulence and emphasize the pleiotropy and adaptability of bacteria occupying diverse niches. IMPORTANCEStaphylococcus saprophyticus is an important cause of urinary tract infections (UTI) in women; such UTI are common, can be severe, and are associated with significant impacts to public health. In addition to being a cause of human UTI, S. saprophyticus can be found in the environment, in food, and associated with animals. After discovering that UTI strains of S. saprophyticus are for the most part closely related to each other, we sought to determine whether these strains are specially adapted to cause disease in humans. We found evidence suggesting that a mutation in the gene aas is advantageous in the context of human infection. We hypothesize that the mutation allows S. saprophyticus to survive better in the human urinary tract. These results show how bacteria found in the environment can evolve to cause disease.
Keywords: Staphylococcus saprophyticus; adhesins; evolution; positive selection; urinary tract infection.
Figures






References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
