Alternative start and termination sites of transcription drive most transcript isoform differences across human tissues
- PMID: 29202200
- PMCID: PMC5778607
- DOI: 10.1093/nar/gkx1165
Alternative start and termination sites of transcription drive most transcript isoform differences across human tissues
Abstract
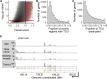
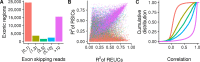
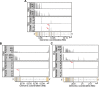
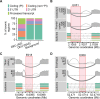
Most human genes generate multiple transcript isoforms. The differential expression of these isoforms can help specify cell types. Diverse transcript isoforms arise from the use of alternative transcription start sites, polyadenylation sites and splice sites; however, the relative contribution of these processes to isoform diversity in normal human physiology is unclear. To address this question, we investigated cell type-dependent differences in exon usage of over 18 000 protein-coding genes in 23 cell types from 798 samples of the Genotype-Tissue Expression Project. We found that about half of the expressed genes displayed tissue-dependent transcript isoforms. Alternative transcription start and termination sites, rather than alternative splicing, accounted for the majority of tissue-dependent exon usage. We confirmed the widespread tissue-dependent use of alternative transcription start sites in a second, independent dataset, Cap Analysis of Gene Expression data from the FANTOM consortium. Moreover, our results indicate that most tissue-dependent splicing involves untranslated exons and therefore may not increase proteome complexity. Thus, alternative transcription start and termination sites are the principal drivers of transcript isoform diversity across tissues, and may underlie the majority of cell type specific proteomes and functions.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- de Klerk E., ’t Hoen P.A.. Alternative mRNA transcription, processing, and translation: insights from RNA sequencing. Trends Genet. 2015; 31:128–139. - PubMed
-
- Breitbart R.E., Andreadis A., Nadal-Ginard B.. Alternative splicing: a ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu. Rev. Biochem. 1987; 56:467–495. - PubMed
-
- Keren H., Lev-Maor G., Ast G.. Alternative splicing and evolution: diversification, exon definition and function. Nat. Rev. Genet. 2010; 11:345–355. - PubMed
-
- Carninci P., Sandelin A., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C.A.M., Taylor M.S., Engström P.G., Frith M.C. et al. . Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006; 38:626–635. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

