CRISPR-Cas9 screen reveals a MYCN-amplified neuroblastoma dependency on EZH2
- PMID: 29202477
- PMCID: PMC5749506
- DOI: 10.1172/JCI90793
CRISPR-Cas9 screen reveals a MYCN-amplified neuroblastoma dependency on EZH2
Abstract
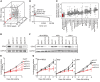
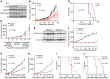
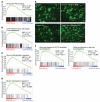
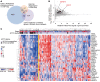
Pharmacologically difficult targets, such as MYC transcription factors, represent a major challenge in cancer therapy. For the childhood cancer neuroblastoma, amplification of the oncogene MYCN is associated with high-risk disease and poor prognosis. Here, we deployed genome-scale CRISPR-Cas9 screening of MYCN-amplified neuroblastoma and found a preferential dependency on genes encoding the polycomb repressive complex 2 (PRC2) components EZH2, EED, and SUZ12. Genetic and pharmacological suppression of EZH2 inhibited neuroblastoma growth in vitro and in vivo. Moreover, compared with neuroblastomas without MYCN amplification, MYCN-amplified neuroblastomas expressed higher levels of EZH2. ChIP analysis showed that MYCN binds at the EZH2 promoter, thereby directly driving expression. Transcriptomic and epigenetic analysis, as well as genetic rescue experiments, revealed that EZH2 represses neuronal differentiation in neuroblastoma in a PRC2-dependent manner. Moreover, MYCN-amplified and high-risk primary tumors from patients with neuroblastoma exhibited strong repression of EZH2-regulated genes. Additionally, overexpression of IGFBP3, a direct EZH2 target, suppressed neuroblastoma growth in vitro and in vivo. We further observed strong synergy between histone deacetylase inhibitors and EZH2 inhibitors. Together, these observations demonstrate that MYCN upregulates EZH2, leading to inactivation of a tumor suppressor program in neuroblastoma, and support testing EZH2 inhibitors in patients with MYCN-amplified neuroblastoma.
Keywords: Epigenetics; Oncology.
Conflict of interest statement
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

