Ligand-activated BMP signaling inhibits cell differentiation and death to promote melanoma
- PMID: 29202482
- PMCID: PMC5749509
- DOI: 10.1172/JCI92513
Ligand-activated BMP signaling inhibits cell differentiation and death to promote melanoma
Abstract
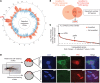
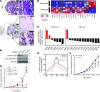
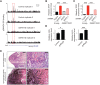
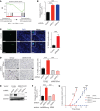
Oncogenomic studies indicate that copy number variation (CNV) alters genes involved in tumor progression; however, identification of specific driver genes affected by CNV has been difficult, as these rearrangements are often contained in large chromosomal intervals among several bystander genes. Here, we addressed this problem and identified a CNV-targeted oncogene by performing comparative oncogenomics of human and zebrafish melanomas. We determined that the gene encoding growth differentiation factor 6 (GDF6), which is the ligand for the BMP family, is recurrently amplified and transcriptionally upregulated in melanoma. GDF6-induced BMP signaling maintained a trunk neural crest gene signature in melanomas. Additionally, GDF6 repressed the melanocyte differentiation gene MITF and the proapoptotic factor SOX9, thereby preventing differentiation, inhibiting cell death, and promoting tumor growth. GDF6 was specifically expressed in melanomas but not melanocytes. Moreover, GDF6 expression levels in melanomas were inversely correlated with patient survival. Our study has identified a fundamental role for GDF6 and BMP signaling in governing an embryonic cell gene signature to promote melanoma progression, thus providing potential opportunities for targeted therapy to treat GDF6-positive cancers.
Keywords: Cancer; Development; Oncology.
Conflict of interest statement
Figures


Similar articles
-
Regulation of zebrafish melanocyte development by ligand-dependent BMP signaling.Elife. 2019 Dec 23;8:e50047. doi: 10.7554/eLife.50047. Elife. 2019. PMID: 31868592 Free PMC article.
-
Ror2 signaling is required for local upregulation of GDF6 and activation of BMP signaling at the neural plate border.Development. 2016 Sep 1;143(17):3182-94. doi: 10.1242/dev.135426. Development. 2016. PMID: 27578181
-
The BMP ligand Gdf6 prevents differentiation of coronal suture mesenchyme in early cranial development.PLoS One. 2012;7(5):e36789. doi: 10.1371/journal.pone.0036789. Epub 2012 May 31. PLoS One. 2012. PMID: 22693558 Free PMC article.
-
[Malignant melanoma and the role of the paradoxal protein Microphthalmia transcription factor].Bull Cancer. 2007 Jan;94(1):81-92. Bull Cancer. 2007. PMID: 17237008 Review. French.
-
Bone morphogenetic proteins.Growth Factors. 2004 Dec;22(4):233-41. doi: 10.1080/08977190412331279890. Growth Factors. 2004. PMID: 15621726 Review.
Cited by
-
SATB2 induction of a neural crest mesenchyme-like program drives melanoma invasion and drug resistance.Elife. 2021 Feb 2;10:e64370. doi: 10.7554/eLife.64370. Elife. 2021. PMID: 33527896 Free PMC article.
-
Long-term non-invasive drug treatments in adult zebrafish that lead to melanoma drug resistance.Dis Model Mech. 2022 May 1;15(5):dmm049401. doi: 10.1242/dmm.049401. Epub 2022 May 9. Dis Model Mech. 2022. PMID: 35394030 Free PMC article.
-
Immune escape and metastasis mechanisms in melanoma: breaking down the dichotomy.Front Immunol. 2024 Feb 14;15:1336023. doi: 10.3389/fimmu.2024.1336023. eCollection 2024. Front Immunol. 2024. PMID: 38426087 Free PMC article. Review.
-
Bone Morphogenic Protein Signaling and Melanoma.Curr Treat Options Oncol. 2021 Apr 17;22(6):48. doi: 10.1007/s11864-021-00849-w. Curr Treat Options Oncol. 2021. PMID: 33866453 Review.
-
Evidence of Cooperation between Hippo Pathway and RAS Mutation in Thyroid Carcinomas.Cancers (Basel). 2021 May 12;13(10):2306. doi: 10.3390/cancers13102306. Cancers (Basel). 2021. PMID: 34065786 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

