Granatum: a graphical single-cell RNA-Seq analysis pipeline for genomics scientists
- PMID: 29202807
- PMCID: PMC5716224
- DOI: 10.1186/s13073-017-0492-3
Granatum: a graphical single-cell RNA-Seq analysis pipeline for genomics scientists
Abstract
Background: Single-cell RNA sequencing (scRNA-Seq) is an increasingly popular platform to study heterogeneity at the single-cell level. Computational methods to process scRNA-Seq data are not very accessible to bench scientists as they require a significant amount of bioinformatic skills.
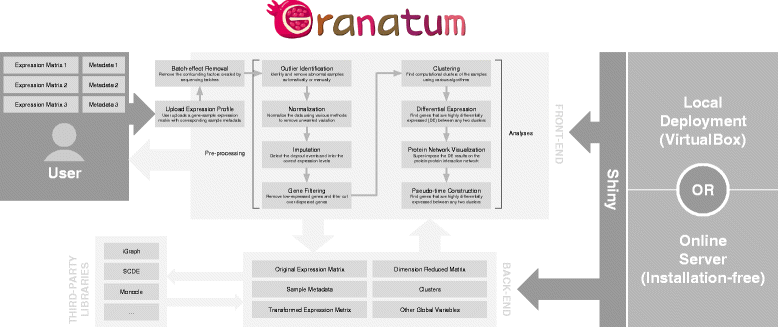
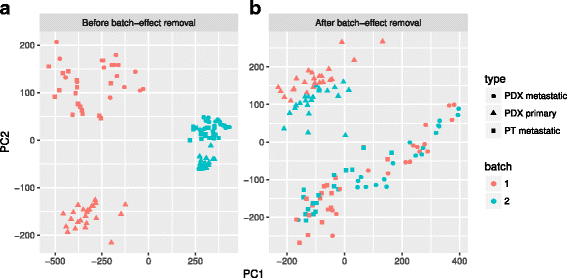
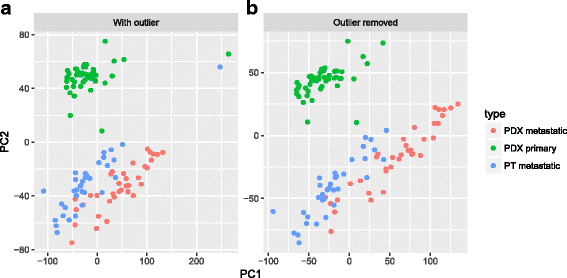
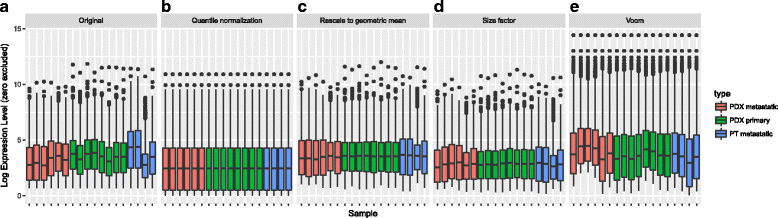
Results: We have developed Granatum, a web-based scRNA-Seq analysis pipeline to make analysis more broadly accessible to researchers. Without a single line of programming code, users can click through the pipeline, setting parameters and visualizing results via the interactive graphical interface. Granatum conveniently walks users through various steps of scRNA-Seq analysis. It has a comprehensive list of modules, including plate merging and batch-effect removal, outlier-sample removal, gene-expression normalization, imputation, gene filtering, cell clustering, differential gene expression analysis, pathway/ontology enrichment analysis, protein network interaction visualization, and pseudo-time cell series construction.
Conclusions: Granatum enables broad adoption of scRNA-Seq technology by empowering bench scientists with an easy-to-use graphical interface for scRNA-Seq data analysis. The package is freely available for research use at http://garmiregroup.org/granatum/app.
Keywords: Clustering; Differential expression; Gene expression; Graphical; Imputation; Normalization; Pathway; Pseudo-time; Single-cell; Software.
Conflict of interest statement
Ethics approval and consent to participate
Not sapplicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
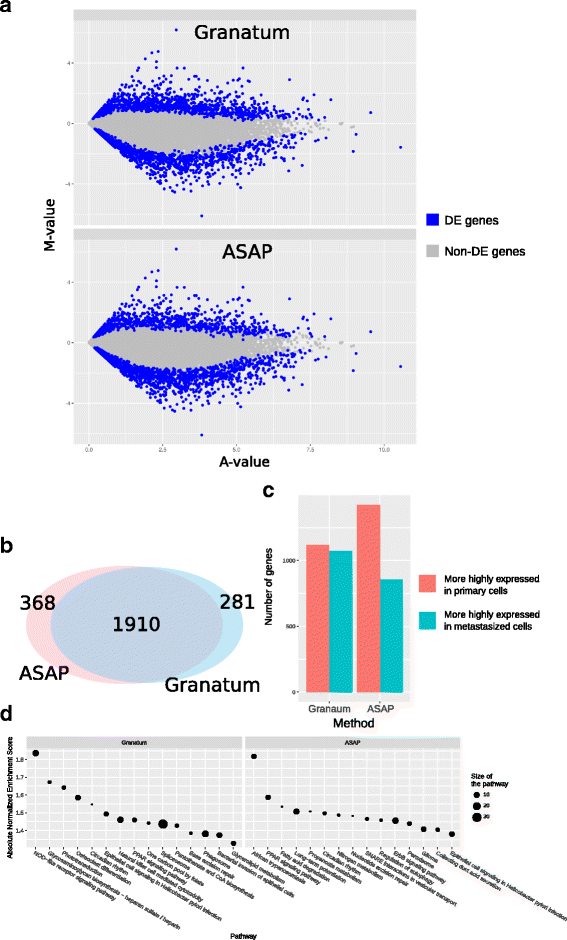
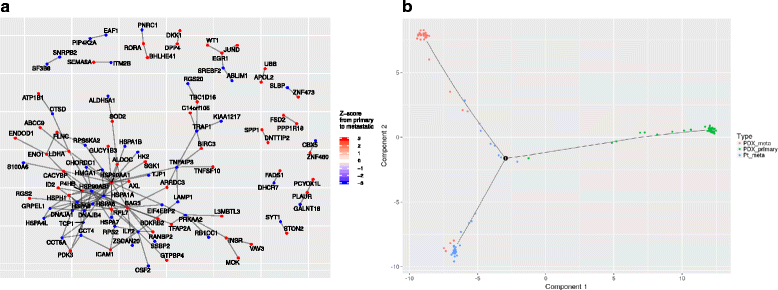
References
-
- Brennecke P, Anders S, Kim JK, Kołodziejczyk AA, Zhang X, Proserpio V, et al. Accounting for technical noise in single-cell RNA-seq experiments. Nat. Methods. 2013;10:1093–5. - PubMed
MeSH terms
Grants and funding
- P20 COBRE GM103457/National Institute of General Medical Sciences (US)
- R01 LM012373/LM/NLM NIH HHS/United States
- R01HD084633/National Institute of Child Health and Human Development (US)
- R01LM012373/U.S. National Library of Medicine
- K01ES025434/National Institute of Environmental Health Sciences (US)
LinkOut - more resources
Full Text Sources
Other Literature Sources

