Experiences from the anatomy track in the ontology alignment evaluation initiative
- PMID: 29202830
- PMCID: PMC5715990
- DOI: 10.1186/s13326-017-0166-5
Experiences from the anatomy track in the ontology alignment evaluation initiative
Abstract
Background: One of the longest running tracks in the Ontology Alignment Evaluation Initiative is the Anatomy track which focuses on aligning two anatomy ontologies. The Anatomy track was started in 2005. In 2005 and 2006 the task in this track was to align the Foundational Model of Anatomy and the OpenGalen Anatomy Model. Since 2007 the ontologies used in the track are the Adult Mouse Anatomy and a part of the NCI Thesaurus. Since 2015 the data in the Anatomy track is also used in the Interactive track of the Ontology Alignment Evaluation Initiative.
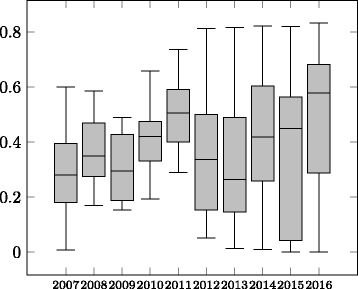
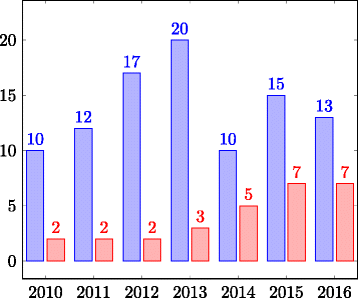
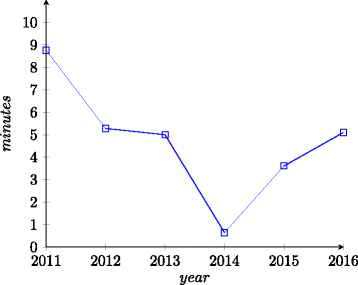
Results: In this paper we focus on the Anatomy track in the years 2007-2016 and the Anatomy part of the Interactive track in 2015-2016. We describe the data set and the changes it went through during the years as well as the challenges it poses for ontology alignment systems. Further, we give an overview of all systems that participated in the track and the techniques they have used. We discuss the performance results of the systems and summarize the general trends.
Conclusions: About 50 systems have participated in the Anatomy track. Many different techniques were used. The most popular matching techniques are string-based strategies and structure-based techniques. Many systems also use auxiliary information. The quality of the alignment has increased for the best performing systems since the beginning of the track and more and more systems check the coherence of the proposed alignment and implement a repair strategy. Further, interacting with an oracle is beneficial.
Keywords: Biomedical ontologies; Ontology alignment; Ontology alignment evaluation initiative.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
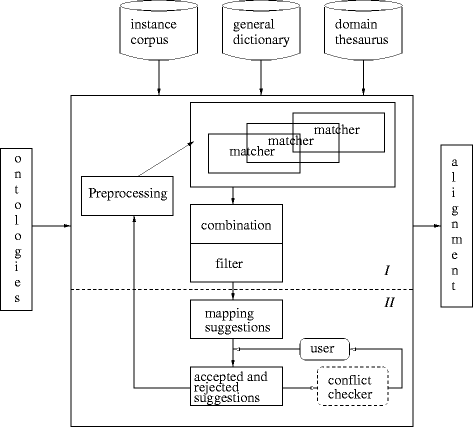
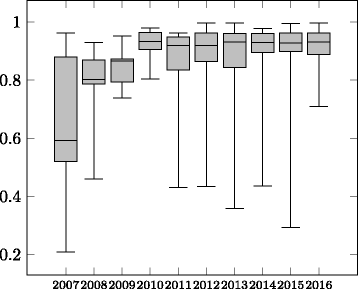
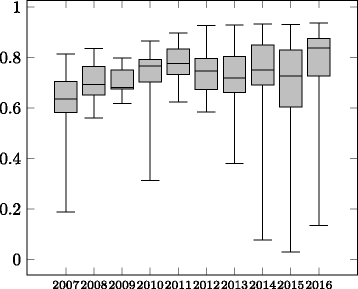
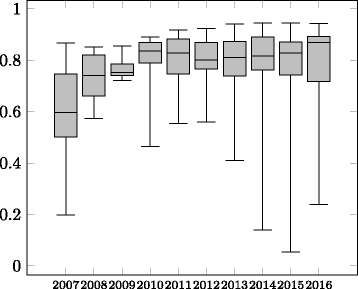
Similar articles
-
Biomedical ontology alignment: an approach based on representation learning.J Biomed Semantics. 2018 Aug 15;9(1):21. doi: 10.1186/s13326-018-0187-8. J Biomed Semantics. 2018. PMID: 30111369 Free PMC article.
-
Interactive biomedical ontology matching.PLoS One. 2019 Apr 17;14(4):e0215147. doi: 10.1371/journal.pone.0215147. eCollection 2019. PLoS One. 2019. PMID: 30995257 Free PMC article.
-
Matching disease and phenotype ontologies in the ontology alignment evaluation initiative.J Biomed Semantics. 2017 Dec 2;8(1):55. doi: 10.1186/s13326-017-0162-9. J Biomed Semantics. 2017. PMID: 29197409 Free PMC article.
-
Ontologies related to livestock for the Global Burden of Animal Diseases programme: a review.Rev Sci Tech. 2024 Aug;43:69-78. doi: 10.20506/rst.43.3519. Rev Sci Tech. 2024. PMID: 39222110 Review. English.
-
The practical impact of ontologies on biomedical informatics.Yearb Med Inform. 2006:124-35. Yearb Med Inform. 2006. PMID: 17051306 Review.
Cited by
-
Integrating ontologies of human diseases, phenotypes, and radiological diagnosis.J Am Med Inform Assoc. 2019 Feb 1;26(2):149-154. doi: 10.1093/jamia/ocy161. J Am Med Inform Assoc. 2019. PMID: 30624645 Free PMC article.
-
Biomedical ontology alignment: an approach based on representation learning.J Biomed Semantics. 2018 Aug 15;9(1):21. doi: 10.1186/s13326-018-0187-8. J Biomed Semantics. 2018. PMID: 30111369 Free PMC article.
-
Variation in the representation of human anatomy within digital resources: Implications for data integration.AMIA Annu Symp Proc. 2018 Dec 5;2018:330-339. eCollection 2018. AMIA Annu Symp Proc. 2018. PMID: 30815072 Free PMC article.
References
-
- Achichi M, Cheatham M, Dragisic Z, Euzenat J, Faria D, Ferrara A, Flouris G, Fundulaki I, Harrow I, Ivanova V, Jiménez-Ruiz E, Kuss E, Lambrix P, Leopold H, Li H, Meilicke C, Montanelli S, Pesquita C, Saveta T, Shvaiko P, Splendiani A, Stuckenschmidt H, Todorov K, Trojahn C, Zamazal O. Results of the ontology alignment evaluation initiative 2016 In: Shvaiko P, Euzenat J, Jiménez-Ruiz E, Cheatham M, Hassanzadeh O, Ichise R, editors. Proceedings of the 11th International Workshop on Ontology Matching, volume 1766 of CEUR Workshop Proceedings: 2016. p. 73–129.
-
- Aguirre JL, Eckert K, Euzenat J, Ferrara A, van Hage WR, Hollink L, Meilicke C, Nikolov A, Ritze D, Scharffe F, Shvaiko P, Sváb-Zamazal O, Trojahn C, Jiménez-Ruiz E, Cuenca Grau B, Zapilko B. Results of the ontology alignment evaluation initiative 2012. In: Shvaiko P, Euzenat J, Kementsietsidis A, Mao M, Noy N, Stuckenschmidt H, editors. Proceedings of the 7th International Workshop on Ontology Matching, volume 946 of CEUR Workshop Proceedings: 2012. p. 73–115.
-
- Ba M, Diallo G. Servomap and servomap-lt results for oaei 2012. In: Shvaiko P, Euzenat J, Kementsietsidis A, Mao M, Noy N, Stuckenschmidt H, editors. Proceedings of the 7th International Workshop on Ontology Matching, volume 946 of CEUR Workshop Proceedings: 2012. p. 197–204.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

