Sensitive detection of HIV-1 resistance to Zidovudine and impact on treatment outcomes in low- to middle-income countries
- PMID: 29202874
- PMCID: PMC5716384
- DOI: 10.1186/s40249-017-0377-0
Sensitive detection of HIV-1 resistance to Zidovudine and impact on treatment outcomes in low- to middle-income countries
Abstract
Background: Thymidine analogs, namely AZT (Zidovudine or Retrovir™) and d4T (Stavudine or Zerit™) are antiretroviral drugs still employed in over 75% of first line combination antiretroviral therapy (cART) in Kampala, Uganda despite aversion to prescribing these drugs for cART in high income countries due in part to adverse events. For this study, we explored how the continued use of these thymidine analogs in cART could impact emergence of drug resistance and impact on future treatment success in Uganda, a low-income country.
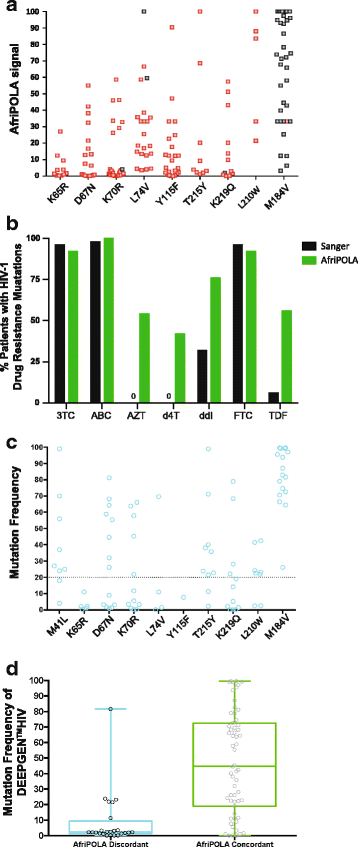
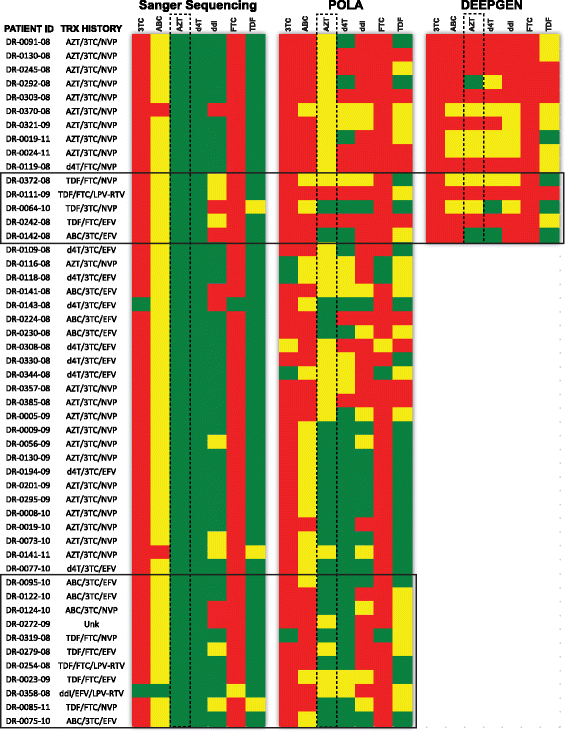
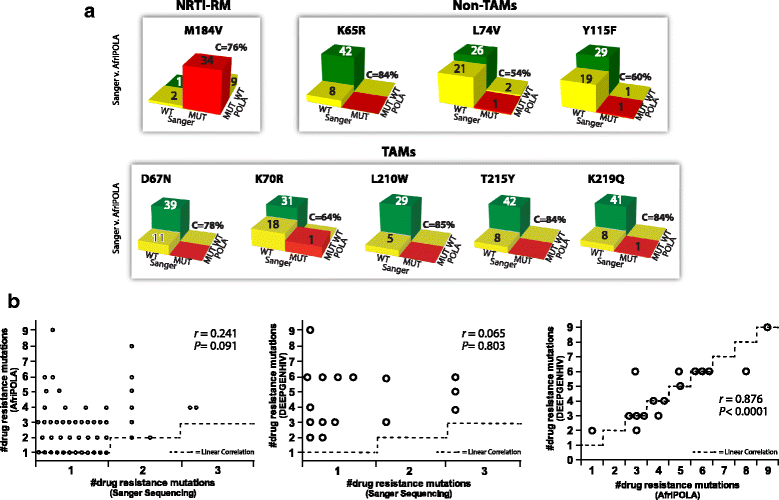
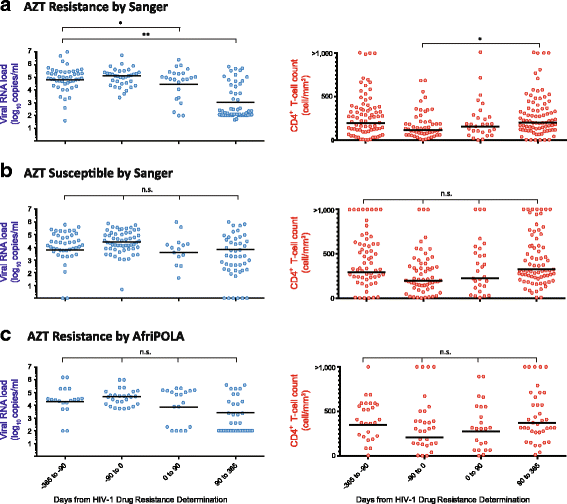
Methods: We examined the drug resistance genotypes by Sanger sequencing of 262 HIV-infected patients failing a first line combined antiretroviral treatment containing either AZT or d4T, which represents approximately 5% of the patients at the Joint Clinical Research Center receiving a AZT or d4T containing treatment. Next generation sequencing (DEEPGEN™HIV) and multiplex oligonucleotide ligation assays (AfriPOLA) were then performed on a subset of patient samples to detect low frequency drug resistant mutations. CD4 cell counts, viral RNA loads, and treatment changes were analyzed in a cohort of treatment success and failures.
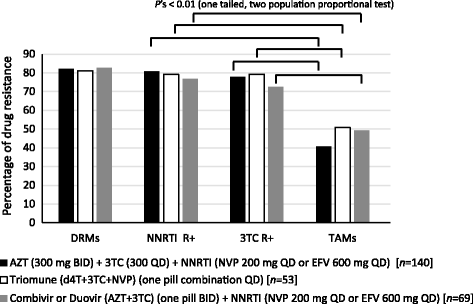
Results: Over 80% of patients failing first line AZT/d4T-containing cART had predicted drug resistance to 3TC (Lamivudine) and non-nucleoside RT inhibitors (NNRTIs) in the treatment regimen but only 45% had resistance AZT/d4T associated resistance mutations (TAMs). TAMs were however detected at low frequency within the patients HIV quasispecies (1-20%) in 21 of 34 individuals who were failing first-line AZT-containing cART and lacked TAMs by Sanger. Due to lack of TAMs by Sanger, AZT was typically maintained in second-line therapies and these patients had a low frequency of subsequent virologic success.
Conclusions: Our findings suggest that continued use of AZT and d4T in first-line treatment in low-to-middle income countries may lead to misdiagnosis of HIV-1 drug resistance and possibly enhance a succession of second- and third-line treatment failures.
Keywords: Antiretroviral treatment; Drug resistance; Uganda.
Conflict of interest statement
Ethics approval and consent to participate
Clinical and virological data were obtained from patient care database at the JCRC under IRB approval (EM10-07) for HIV-1 drug resistance testing.
Consent for publication
Not applicable
Competing interests
E.J.A developed and patented the technology used in AfriPOLA. R.M.G and M.E.Q-M developed DEEPGEN™HIV. There are no further patents, products in development or marketed products to declare.
Figures
References
-
- UNAIDS. The HIV and AIDS Uganda Country Progress Report 2014. Geneva, Switzerland. 2014; http://www.unaids.org/sites/default/files/country/documents/UGA_narrativ....
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

