The generation of a simian adenoviral vectored HCV vaccine encoding genetically conserved gene segments to target multiple HCV genotypes
- PMID: 29203182
- PMCID: PMC5756538
- DOI: 10.1016/j.vaccine.2017.10.079
The generation of a simian adenoviral vectored HCV vaccine encoding genetically conserved gene segments to target multiple HCV genotypes
Abstract
Background: Hepatitis C virus (HCV) genomic variability is a major challenge to the generation of a prophylactic vaccine. We have previously shown that HCV specific T-cell responses induced by a potent T-cell vaccine encoding a single strain subtype-1b immunogen target epitopes dominant in natural infection. However, corresponding viral regions are highly variable at a population level, with a reduction in T-cell reactivity to these variants. We therefore designed and manufactured second generation simian adenovirus vaccines encoding genomic segments, conserved between viral genotypes and assessed these for immunogenicity.
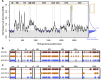
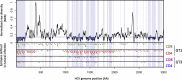
Methods: We developed a computer algorithm to identify HCV genomic regions that were conserved between viral subtypes. Conserved segments below a pre-defined diversity threshold spanning the entire HCV genome were combined to create novel immunogens (1000-1500 amino-acids), covering variation in HCV subtypes 1a and 1b, genotypes 1 and 3, and genotypes 1-6 inclusive. Simian adenoviral vaccine vectors (ChAdOx) encoding HCV conserved immunogens were constructed. Immunogenicity was evaluated in C57BL6 mice using panels of genotype-specific peptide pools in ex-vivo IFN-ϒ ELISpot and intracellular cytokine assays.
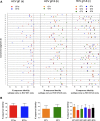
Results: ChAdOx1 conserved segment HCV vaccines primed high-magnitude, broad, cross-reactive T-cell responses; the mean magnitude of total HCV specific T-cell responses was 1174 SFU/106 splenocytes for ChAdOx1-GT1-6 in C57BL6 mice targeting multiple genomic regions, with mean responses of 935, 1474 and 1112 SFU/106 against genotype 1a, 1b and 3a peptide panels, respectively. Functional assays demonstrated IFNg and TNFa production by vaccine-induced CD4 and CD8 T-cells. In silico analysis shows that conserved immunogens contain multiple epitopes, with many described in natural HCV infection, predicting immunogenicity in humans.
Conclusions: Simian adenoviral vectored vaccines encoding genetic segments that are conserved between all major HCV genotypes contain multiple T-cell epitopes and are highly immunogenic in pre-clinical models. These studies pave the way for the assessment of multi-genotypic HCV T-cell vaccines in humans.
Keywords: Conserved segments; Cross-reactivity; HCV vaccine; Simian adenovirus; T cell.
Copyright © 2017. Published by Elsevier Ltd.
Figures

References
-
- Foster G.R. Sofosbuvir and velpatasvir for HCV genotype 2 and 3 infection. N Engl J Med. 2015;373:2608–2617. - PubMed
-
- Feld J.J. Sofosbuvir and velpatasvir for HCV genotype 1, 2, 4, 5, and 6 infection. N Engl J Med. 2015;373:2599–2607. - PubMed
-
- Cammà C. Cost-effectiveness of boceprevir or telaprevir for previously treated patients with genotype 1 chronic hepatitis C. J Hepatol. 2013;59:658–666. - PubMed
-
- Volk M.L. Antiviral therapy for hepatitis C: why are so few patients being treated? J Antimicrob Chemother. 2010;65:1327–1329. - PubMed
-
- Razavi H., Estes C., Pasini K., Gower E., Hindman S. HCV treatment rate in select european countries in 2004–2010. J Hepatol Supplement. 2013;1:S22–S23.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

