Extended Linkers Improve the Detection of Protein-protein Interactions (PPIs) by Dihydrofolate Reductase Protein-fragment Complementation Assay (DHFR PCA) in Living Cells
- PMID: 29203496
- PMCID: PMC5795398
- DOI: 10.1074/mcp.TIR117.000385
Extended Linkers Improve the Detection of Protein-protein Interactions (PPIs) by Dihydrofolate Reductase Protein-fragment Complementation Assay (DHFR PCA) in Living Cells
Erratum in
-
Extended linkers improve the detection of protein-protein interactions (PPIs) by dihydrofolate reductase protein-fragment complementation assay (DHFR PCA) in living cells.Mol Cell Proteomics. 2018 Mar;17(3):549. doi: 10.1074/mcp.A117.000385. Mol Cell Proteomics. 2018. PMID: 29496912 Free PMC article. No abstract available.
Abstract
Understanding the function of cellular systems requires describing how proteins assemble with each other into transient and stable complexes and to determine their spatial relationships. Among the tools available to perform these analyses on a large scale is Protein-fragment Complementation Assay based on the dihydrofolate reductase (DHFR PCA). Here we test how longer linkers between the fusion proteins and the reporter fragments affect the performance of this assay. We investigate the architecture of the RNA polymerases, the proteasome and the conserved oligomeric Golgi (COG) complexes in living cells and performed large-scale screens with these extended linkers. We show that longer linkers significantly improve the detection of protein-protein interactions and allow to measure interactions further in space than the standard ones. We identify new interactions, for instance between the retromer complex and proteins related to autophagy and endocytosis. Longer linkers thus contribute an enhanced additional tool to the existing toolsets for the detection and measurements of protein-protein interactions and protein proximity in living cells.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
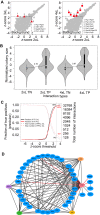
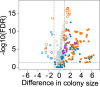
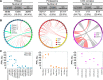

References
-
- Diss G., Dube A. K., Boutin J., Gagnon-Arsenault I., and Landry C. R. (2013) A systematic approach for the genetic dissection of protein complexes in living cells. Cell Rep. 3, 2155–2167 - PubMed
-
- Gagnon-Arsenault I., Marois Blanchet F. C., Rochette S., Diss G., Dube A. K., and Landry C. R. (2013) Transcriptional divergence plays a role in the rewiring of protein interaction networks after gene duplication. J. Proteomics 81, 112–125 - PubMed
-
- Vo T. V., Das J., Meyer M. J., Cordero N. A., Akturk N., Wei X., Fair B. J., Degatano A. G., Fragoza R., Liu L. G., Matsuyama A., Trickey M., Horibata S., Grimson A., Yamano H., Yoshida M., Roth F. P., Pleiss J. A., Xia Y., and Yu H. (2016) A Proteome-wide Fission Yeast Interactome Reveals Network Evolution Principles from Yeasts to Human. Cell 164, 310–323 - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

