Novel and shared neoantigen derived from histone 3 variant H3.3K27M mutation for glioma T cell therapy
- PMID: 29203539
- PMCID: PMC5748856
- DOI: 10.1084/jem.20171046
Novel and shared neoantigen derived from histone 3 variant H3.3K27M mutation for glioma T cell therapy
Abstract
The median overall survival for children with diffuse intrinsic pontine glioma (DIPG) is less than one year. The majority of diffuse midline gliomas, including more than 70% of DIPGs, harbor an amino acid substitution from lysine (K) to methionine (M) at position 27 of histone 3 variant 3 (H3.3). From a CD8+ T cell clone established by stimulation of HLA-A2+ CD8+ T cells with synthetic peptide encompassing the H3.3K27M mutation, complementary DNA for T cell receptor (TCR) α- and β-chains were cloned into a retroviral vector. TCR-transduced HLA-A2+ T cells efficiently killed HLA-A2+H3.3K27M+ glioma cells in an antigen- and HLA-specific manner. Adoptive transfer of TCR-transduced T cells significantly suppressed the progression of glioma xenografts in mice. Alanine-scanning assays suggested the absence of known human proteins sharing the key amino acid residues required for recognition by the TCR, suggesting that the TCR could be safely used in patients. These data provide us with a strong basis for developing T cell-based therapy targeting this shared neoepitope.
© 2018 Chheda et al.
Figures
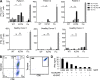


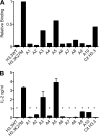


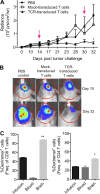
Comment in
-
Shared cancer neoantigens: Making private matters public.J Exp Med. 2018 Jan 2;215(1):5-7. doi: 10.1084/jem.20172188. Epub 2017 Dec 21. J Exp Med. 2018. PMID: 29269561 Free PMC article.
References
-
- Adams J.J., Narayanan S., Birnbaum M.E., Sidhu S.S., Blevins S.J., Gee M.H., Sibener L.V., Baker B.M., Kranz D.M., and Garcia K.C.. 2016. Structural interplay between germline interactions and adaptive recognition determines the bandwidth of TCR-peptide-MHC cross-reactivity. Nat. Immunol. 17:87–94. 10.1038/ni.3310 - DOI - PMC - PubMed
-
- Bakker A.B., Marland G., de Boer A.J., Huijbens R.J., Danen E.H., Adema G.J., and Figdor C.G.. 1995. Generation of antimelanoma cytotoxic T lymphocytes from healthy donors after presentation of melanoma-associated antigen-derived epitopes by dendritic cells in vitro. Cancer Res. 55:5330–5334. - PubMed
-
- Bassani-Sternberg M., Pletscher-Frankild S., Jensen L.J., and Mann M.. 2015. Mass spectrometry of human leukocyte antigen class I peptidomes reveals strong effects of protein abundance and turnover on antigen presentation. Mol. Cell. Proteomics. 14:658–673. 10.1074/mcp.M114.042812 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

