Bayesian analysis of genome-wide inflammatory bowel disease data sets reveals new risk loci
- PMID: 29203833
- PMCID: PMC5838995
- DOI: 10.1038/s41431-017-0041-y
Bayesian analysis of genome-wide inflammatory bowel disease data sets reveals new risk loci
Abstract
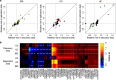
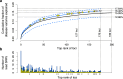
Genome-wide association studies (GWAS) have identified and validated 200 risk loci for inflammatory bowel disease (IBD) to date, including risk loci for both Crohn's disease and ulcerative colitis. Previously studies mainly used single SNP testing methods and only reported the most significant association within each locus. Advanced methods are needed to detect additional joint effects of multiple SNPs and fine map causal variants in presence of strong linkage disequilibrium. In this study, we applied a powerful Bayesian method to analyze an existing Immunochip data sets for IBD from the international inflammatory bowel disease genetics consortium. The method jointly tested single and set-based SNPs in a unified framework and filtered indirect associations due to linkage disequilibrium, thereby fine-mapping the most likely IBD variants. Using an independent collection of individuals from 11 IBD GWAS as validation, our approach discovered and validated 9 completely new IBD loci and 5 independent signals (excluding the major histocompatibility complex) near known IBD loci reaching genome-wide significance. Several of the replicated new loci implicated functionally more interpretable genes than previous reports. The epigenetic marks at our detected IBD signals demonstrated significant activation signatures in blood cell types and correspondingly substantial repression in stem cells, suggesting regulatory links between genetic variants and IBD. Our analysis of the currently largest IBD datasets therefore added new insights that will be integral to the ongoing efforts in IBD genetics.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

