TET proteins safeguard bivalent promoters from de novo methylation in human embryonic stem cells
- PMID: 29203910
- PMCID: PMC5742051
- DOI: 10.1038/s41588-017-0002-y
TET proteins safeguard bivalent promoters from de novo methylation in human embryonic stem cells
Erratum in
-
Author Correction: TET proteins safeguard bivalent promoters from de novo methylation in human embryonic stem cells.Nat Genet. 2018 May;50(5):764. doi: 10.1038/s41588-017-0025-4. Nat Genet. 2018. PMID: 29255265
-
Publisher Correction: TET proteins safeguard bivalent promoters from de novo methylation in human embryonic stem cells.Nat Genet. 2018 May;50(5):764. doi: 10.1038/s41588-017-0024-5. Nat Genet. 2018. PMID: 29255266
Abstract
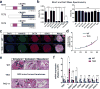
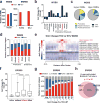
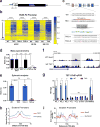
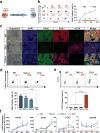
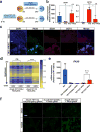
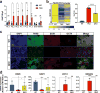
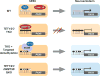
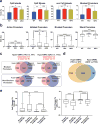
TET enzymes oxidize 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), which can lead to DNA demethylation. However, direct connections between TET-mediated DNA demethylation and transcriptional output are difficult to establish owing to challenges in distinguishing global versus locus-specific effects. Here we show that TET1, TET2 and TET3 triple-knockout (TKO) human embryonic stem cells (hESCs) exhibit prominent bivalent promoter hypermethylation without an overall corresponding decrease in gene expression in the undifferentiated state. Focusing on the bivalent PAX6 locus, we find that increased DNMT3B binding is associated with promoter hypermethylation, which precipitates a neural differentiation defect and failure of PAX6 induction during differentiation. dCas9-mediated locus-specific demethylation and global inactivation of DNMT3B in TKO hESCs partially reverses the hypermethylation at the PAX6 promoter and improves differentiation to neuroectoderm. Taking these findings together with further genome-wide methylation and TET1 and DNMT3B ChIP-seq analyses, we conclude that TET proteins safeguard bivalent promoters from de novo methylation to ensure robust lineage-specific transcription upon differentiation.
Conflict of interest statement
5hmC-Seal has been licensed to Active Motif and Epican by the University of Chicago.
Figures
Similar articles
-
TET1 Deficiency Impairs Morphogen-free Differentiation of Human Embryonic Stem Cells to Neuroectoderm.Sci Rep. 2020 Jun 25;10(1):10343. doi: 10.1038/s41598-020-67143-x. Sci Rep. 2020. PMID: 32587369 Free PMC article.
-
Distinct and overlapping control of 5-methylcytosine and 5-hydroxymethylcytosine by the TET proteins in human cancer cells.Genome Biol. 2014 Jun 23;15(6):R81. doi: 10.1186/gb-2014-15-6-r81. Genome Biol. 2014. PMID: 24958354 Free PMC article.
-
Deletion of Tet proteins results in quantitative disparities during ESC differentiation partially attributable to alterations in gene expression.BMC Dev Biol. 2019 Jul 8;19(1):16. doi: 10.1186/s12861-019-0196-6. BMC Dev Biol. 2019. PMID: 31286885 Free PMC article.
-
Mechanisms that regulate the activities of TET proteins.Cell Mol Life Sci. 2022 Jun 15;79(7):363. doi: 10.1007/s00018-022-04396-x. Cell Mol Life Sci. 2022. PMID: 35705880 Free PMC article. Review.
-
TET proteins and 5-methylcytosine oxidation in hematological cancers.Immunol Rev. 2015 Jan;263(1):6-21. doi: 10.1111/imr.12239. Immunol Rev. 2015. PMID: 25510268 Free PMC article. Review.
Cited by
-
Imprecise DNMT1 activity coupled with neighbor-guided correction enables robust yet flexible epigenetic inheritance.Nat Genet. 2020 Aug;52(8):828-839. doi: 10.1038/s41588-020-0661-y. Epub 2020 Jul 20. Nat Genet. 2020. PMID: 32690947
-
Insights into epigenetic patterns in mammalian early embryos.Protein Cell. 2021 Jan;12(1):7-28. doi: 10.1007/s13238-020-00757-z. Epub 2020 Jul 15. Protein Cell. 2021. PMID: 32671792 Free PMC article. Review.
-
Targeting Epigenetic Mechanisms to Treat Alcohol Use Disorders (AUD).Curr Pharm Des. 2021 Oct 5;27(30):3252-3272. doi: 10.2174/1381612827666210203142539. Curr Pharm Des. 2021. PMID: 33535943 Free PMC article.
-
p21 Promoter Methylation Is Vital for the Anticancer Activity of Withaferin A.Int J Mol Sci. 2025 Jan 30;26(3):1210. doi: 10.3390/ijms26031210. Int J Mol Sci. 2025. PMID: 39940977 Free PMC article.
-
Ten-Eleven Translocation Family Proteins: Structure, Biological Functions, Diseases, and Targeted Therapy.MedComm (2020). 2025 Jul 1;6(7):e70245. doi: 10.1002/mco2.70245. eCollection 2025 Jul. MedComm (2020). 2025. PMID: 40599235 Free PMC article. Review.
References
-
- Robertson KD. DNA methylation and human disease. Nature Reviews Genetics. 2005;6:597–610. - PubMed
-
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. - PubMed
-
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. - PubMed
-
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nature Reviews Genetics. 2013;14:204–220. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

